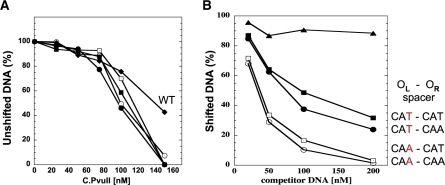
Figure 4.
In vitro interaction of C.PvuII protein with wild-type and altered C-box regions. (A) EMSA reactions were processed as outlined in ‘Materials and methods’ section and shown in Supplementary Data (Figure S4). Data are shown as % of unshifted DNA versus increasing concentration of added C.PvuII. ‘WT’ refers to the native PvuII C-boxes and spacers. The others all have symmetrized spacers with varied intra-operator spacers. (B) EMSA competition assays were performed using 200 nM of C.PvuII and 20 nM of biotin-labeled WT C-box 126-mer (as described in ‘Materials and methods’ section). Competition reactions contained increasing amounts of unlabeled 126-mer DNA fragments (from 1- to 10-fold molar excess). The competitor DNAs contained intra-operator spacers: CAA/CAA (white circles), CAA/CAT (white squares), CAT/CAA (black squares), CAT/CAT (black circles) or no C-boxes (triangles, negative control). Following EMSA and electroblotting, the shifted bands for each reaction were visualized and quantified via chemiluminescent detection of the biotinylated DNA as described in ‘Materials and methods’ section. For clarity symbols are the same as in Figure 3.