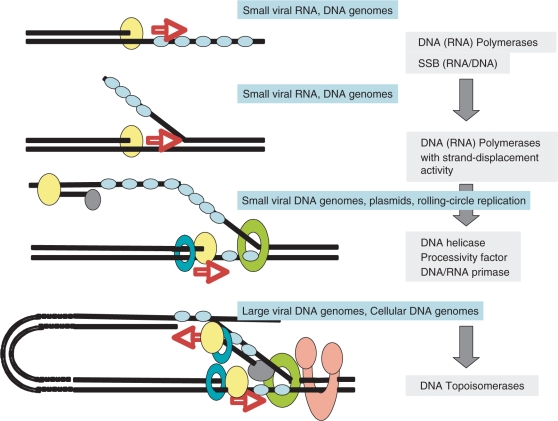
Figure 3.
Hypothetical and schematic scenario for the evolution of the elongation step of RNA and DNA replication, from simple to complex, with the progressive ‘invention’ of enzymes specifically involved in genome replication. Steps 1 and 2, asymetric replication (one strand at a time), observed in organisms with single or double-stranded RNA or DNA genomes. A complementary strand is first synthesized and will serve of template for the synthesis of the new strand (step 2). These steps require more and more processive polymerases and single-stranded DNA or RNA-binding proteins. Step 3, partially symmetric replication (one strand starts to be replicated before the first one has been fully replicated). This introduces the notion of leading and lagging strands and requires the recruitment of a primase activity possibly previously only used in the initiation step (together with other mechanisms such as tRNA priming and protein priming). Steps 2 and 3 can be progressively improved by the introduction of helicases and processivity factors (clamp-like) to help the polymerase. Step 4, symetric replication with two polymerases and the primase activity linked to the helicase activity. The formation of short fragments on the lagging strand require the intervention of other proteins (nuclease/ligases) which have been omitted for clarity. This step can be improved by coupling the two polymerases in a physical complex and by rotating the lagging strand by 180° to allow concurrent replication of the two strands. At this stage, topoisomerases are required to replicate long linear genomes or circular genomes. All steps in that scenario are observed today in the viral world. Steps 1 and 2 in both RNA and DNA viruses, step 3 only in DNA viruses and step 4 in both DNA viruses with large genomes (including concurrent replication) and in cellular organisms.