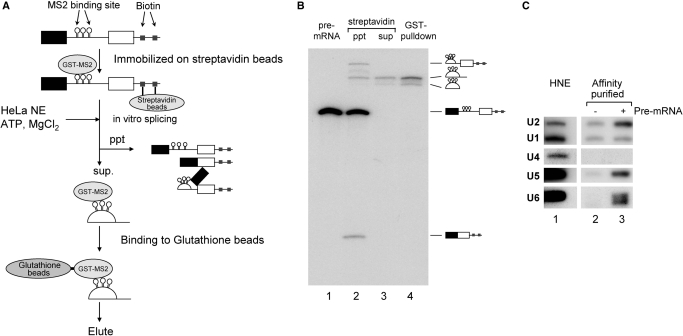
Figure 1.
Purification of the post-splicing intron complex from an in vitro splicing reaction. (A) A schematic representation of the isolation of the intron RNP complex from splicing reaction mixture. Chicken c-crystallin splicing substrate with three MS2 binding loops in its intron (δ-3MS2) was ligated with an oligonucleotide DNA containing biotinylated linker before being immobilized on streptavidin magnetic beads and bound with recombinant GST-MS2 proteins. After immobilization, this pre-mRNA was spliced in HeLa cell nuclear extracts. The reaction was subjected to precipitation using a magnetic stand to remove the pre-mRNA, splicing intermediates and mRNA. The supernatant was subjected to glutathione beads to purify the intron-RNP, which was then eluted with Sarkosyl-containing elution buffer. (B) Analysis of the RNAs recovered from each step of the intron–RNP purification shown in (A). The RNAs were analyzed by 6% denaturing polyacrylamide gel electrophoresis. The structure of each RNA is schematically shown on the right side of the panel. (C) Northern blotting analysis of the spliceosomal U snRNAs in the isolated intron–RNP. HeLa cell nuclear extracts (HNE; lane 1) that contained all five U snRNAs were used as a positive control. The isolated intron–RNPs with (lane 3) or without (lane 2) exogenously added pre-mRNAs were subjected to northern blotting analysis and probed for the five U snRNAs.