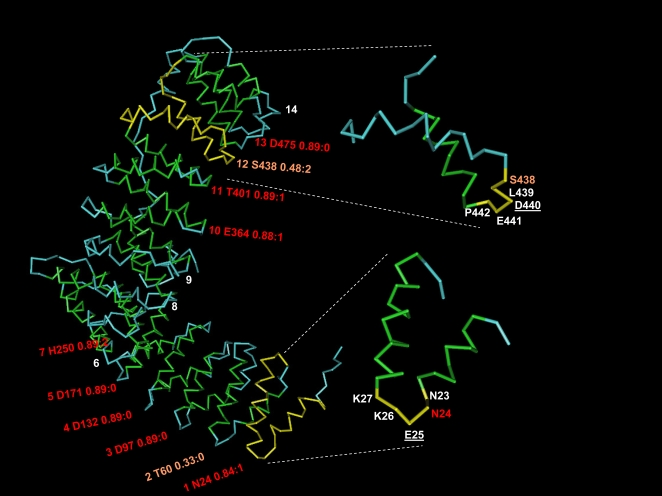
Figure 1. Detection of repeats in an alpha-rod protein.
Structure (alpha-backbone trace) of the 591 aa N-terminal fragment of human adaptor-related protein complex 2, beta 1 subunit, as forming part of the AP2 clathrin adaptor core [69] (PDB code 2VGL chain B). Green and blue represent residues in alpha-helix and in disordered conformation, respectively. This structure has no residue in beta-strand conformation and is entirely composed of an alpha-rod of 14 repeats previously classified as HEAT repeats of type ADB [4]. The label for each repeat indicates the following: repeat order, residue detected by the network, score of hit, and position relative to residue used for training. For example, “1 N24 0.84∶1” indicates that the residue detected for repeat #1 was N (amino acid code for asparagine) in position 24 of the sequence, with score 0.84, but that the residue in relative position 1 (that is, at 25) was the one used to train the network as being in the hinge. Ten out of the 14 repeats were detected, 8 of them with score> = 0.80. The inset shows repeats 12 (right, top) and 1 (right, bottom) with the residue used as positive in the training underscored. A coloured label indicates the residue identified by the network after training, which in both cases is not the one given in the training but others belonging to the hinge (E25 and S438). The figure was generated using NCBI's linked viewer, Cn3D [70].