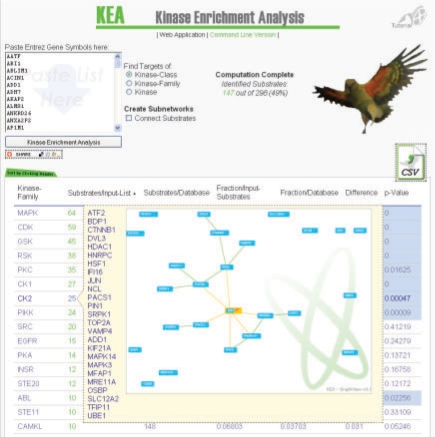
Fig. 1.
Screenshot of the KEA user interface. Users can paste lists of Entrez gene symbols, representing human proteins; select the level of analysis: kinase-class, kinase-family or kinase and then the program outputs a list of ranked kinase-classes, kinase-families or kinases based on specificity of phosphorylating substrates from the input list. Substrates can be then connected based on their known protein–protein interaction using an original network viewer developed using Adobe Flash CS4.