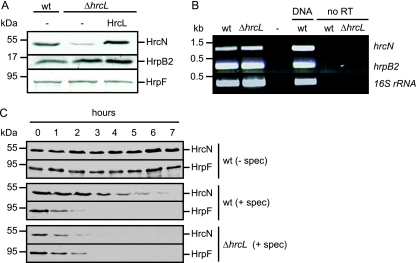
FIG. 5.
HrcL contributes to HrcN stability. (A) HrcN protein amounts are reduced in the absence of HrcL. X. campestris pv. vesicatoria strains 85* (wild type [wt]) and 85*ΔhrcL (ΔhrcL) carrying the empty vector (−) or synthesizing HrcL-c-Myc as indicated were grown in minimal medium A supplemented with sucrose and Casamino Acids. Total protein extracts were analyzed by immunoblotting using HrcN-, HrpB2-, and HrpF-specific antibodies. (B) Transcription of hrcN is not affected in an hrcL deletion mutant. RNA was isolated from X. campestris pv. vesicatoria strains 85* (wt) and 85*ΔhrcL (ΔhrcL). RT-PCR was performed using cDNA and primers specific for hrcN, hrpB2, and the 16S rRNA, which was amplified as a constitutive control. As a positive control for the PCR, we used genomic DNA of strain 85* (DNA). As negative controls, the template was replaced by water (−) or the reaction was performed in the absence of RT (no RT). Amplicons were separated on a 2% agarose gel and stained with ethidium bromide. (C) Protein stability of HrcN is decreased in an hrcL deletion mutant. X. campestris pv. vesicatoria strains 85* (wt) and 85*ΔhrcL (ΔhrcL) were grown in minimal medium A supplemented with sucrose and Casamino Acids. Bacterial cultures were adjusted to an optical density at 600 nm of 0.2 and incubated at 30°C in the absence or presence of spectinomycin (spec) at a final concentration of 400 μg/ml. Samples were taken over a period of 7 h after addition of spectinomycin. Equal amounts of total protein extracts were analyzed by immunoblotting using HrcN- and HrpF-specific antibodies, respectively.