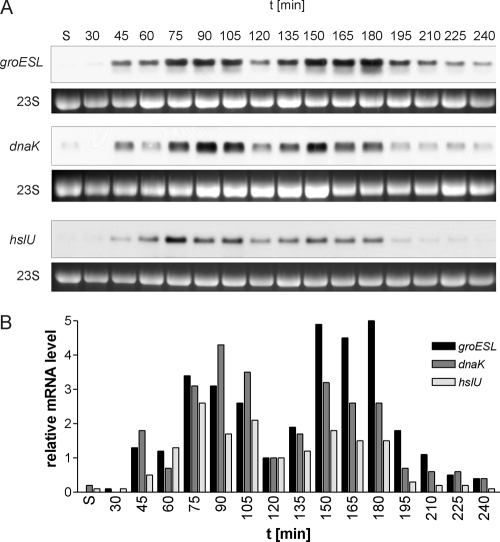
FIG. 4.
Growth cycle-dependent oscillation of mRNA levels of σ32-controlled genes in E. coli MG1655. (A) Total RNA was isolated at the given time points and analyzed by Northern blotting. groESL, dnaK, and hslU mRNAs show similar fluctuation patterns during the course of the growth experiment. Transcript levels are low or below the detection limit in stationary phase, and an increase in the abundance of each transcript can be observed at 45 min, with a first peak at 75 to 90 min. After a decline with a minimum at 120 min, concomitant with the completion of the first round of cell division by most cells in the culture, a second peak appears at 150 min. The transition to stationary phase and the following reduction in cell division activity are accompanied by a marked reduction in transcript levels. 23S rRNA from the ethidium bromide-stained agarose gel is shown as a loading control. (B) Graphical representation of quantitated mRNA levels, showing the oscillation of the levels of σ32-dependent transcripts. The level of each mRNA at 120 min was set to 1. Experiments performed at least in triplicate produced similar results; one representative result is shown. S, stationary phase.