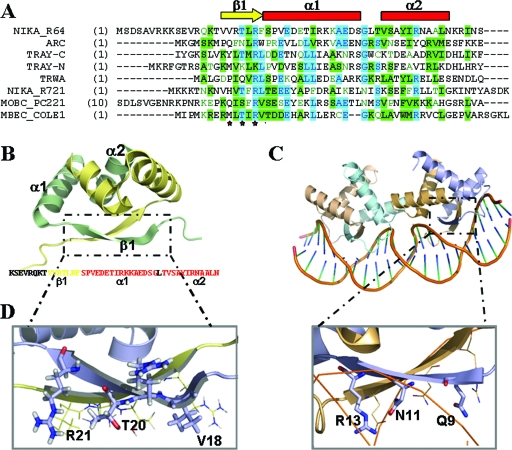
FIG. 5.
(A) CLUSTALW alignment of the 50 N-terminal amino acids of MbeC with other RHH accessory proteins (TrwA_R388, TraY_F [N-terminal and C-terminal domain], NikA_R721, NikA_R64, and MobC_pC221) and the RHH repressor Arc. The secondary structure of the solved NikA_R64 NMR structure is shown above the alignment (the β-sheet is represented by a yellow arrow, and α-helices are indicated by red squares). (B) Ribbon diagram representation of the NMR structure of NikA_R64 N-terminal domain (PDB accession no. 2ba3). One monomer is shown in blue, and the other is shown in wheat color. The sequence of the solved domain is shown below the structure with a color code secondary structure (yellow β-sheet and red α-helix forming residues). (C) Ribbon diagram representation of the crystal structure of two Arc dimers bound to its operator (PDB accession no. 1par). (D) Comparison of NikA (left) and Arc (right) β-sheet polar residues.