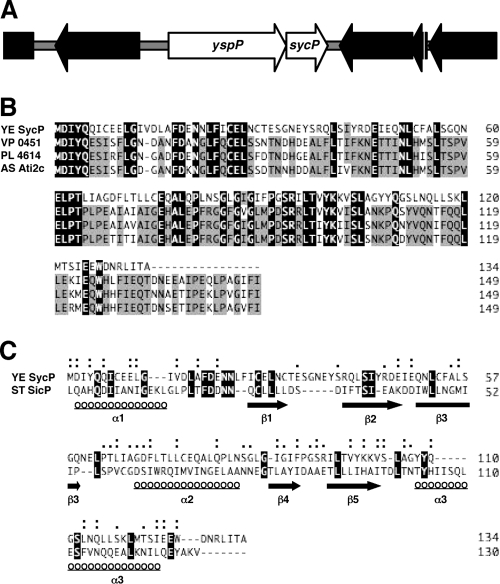
FIG. 1.
(A) Gene organization of the yspP and sycP region. ORFs of yspP and sycP are depicted as white arrows. (B) Alignment of the SycP sequence (YE SycP) with those of Vibrio parahaemolyticus VPA0451 (VP 0451), Photorhabdus luminescens subsp. laumondii plu4614 (PL 4614), and Aeromonas salmonicida subsp. salmonicida ati2 chaperone (AS Ati2c). Identical and conserved (75%) amino acids are highlighted in black and gray, respectively. (C) SicP structure-based alignment of the SycP sequence. Structural homologues were searched by a fold recognition program, phyre (http://www.sbg.bio.ic.ac.uk/phyre/), and the obtained alignment was analyzed by another fold recognition program, FUGUE (http://tardis.nibio.go.jp/fugue/). ST SicP denotes the sequence of Salmonella enterica serovar Typhimurium SicP. Identical amino acids are highlighted in black. Similar and very similar amino acids are marked by single and double dots, respectively. The locations of secondary structures, α-helix and β-sheet, are shown with underlined circles and solid arrows, respectively.