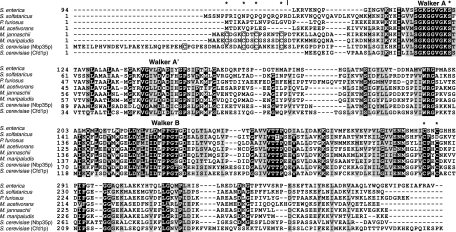
FIG. 1.
A protein sequence alignment of bacterial, archaeal, and eukaryotic ApbC/Nbp35 homologs was constructed using the ClustalW program (version 1.83) (37). The sequence of the S. enterica serovar Typhimurium protein (ApbC; RefSeq accession no. NP_461098.1) is shown without the amino-terminal domain that is not homologous to the amino-terminal domains of the archaeal and eukaryotic proteins. The archaeal homologs are from S. solfataricus (SSO0460; accession no. NP_341994.1), P. furiosus (PF1145; accession no. NP_578874.1), Methanosarcina acetivorans (MA4246; accession no. NP_619111.1), M. jannaschii (MJ0283; accession no. NP_247256.1), and M. maripaludis (MMP0704; accession no. NP_987824.1). The two paralogs from S. cerevisiae are Nbp35 (accession no. NP_011424.1) and Cfd1 (accession no. NP_012263.1). Conserved amino acid residues are shown in white on a black background. Similar residues are shown in black on a gray background. The four conserved amino-terminal cysteine residues shared by the MMP0704 and Nbp35p proteins are boxed. Asterisks above the sequences indicate MMP0704 residues replaced by mutagenesis in this study. A vertical bar indicates the N termini of the truncated proteins MJ0283(19-290) and MMP0704(20-289).