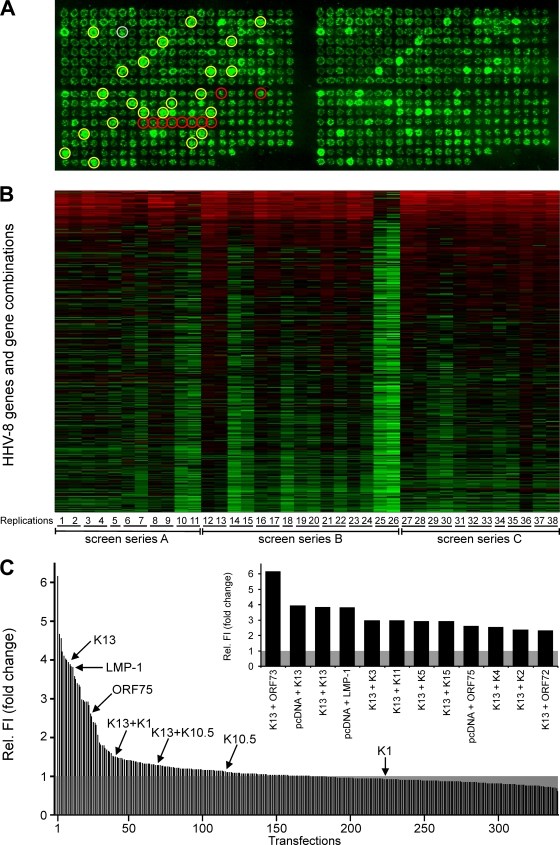
FIG. 3.
Single-gene and combination effects of HHV-8 genes on NF-κB activation. (A) Image of a representative RTCM chip with 371 transfections in duplicate (left and right). Expression plasmids for all HHV-8 genes individually (n = 86) and, additionally, all K and latent genes (19 K genes, ORF72, and ORF73) in all possible pairwise combinations (n = 231) were spotted onto the slide. A plasmid encoding LMP-1 was used as a positive control (gray circle; n = 1) and empty-vector pcDNA (red circles; n = 10) as a negative control. Moreover, the LMP-1 gene was combined with the K genes, ORF72, and ORF73 (n = 21). The reporter plasmid pNF-κB-EGFP was added to all of the described settings. A constitutively GFP-expressing plasmid (pFRED) was spotted at several different positions in order to control transfection efficiency on the chip and to provide a positional marker (yellow circles; n = 22). HEK 293T cells were seeded on RTCM chips. GFP fluorescence was determined with a laser scanner (473 nm) after 48 h. (B) Heat map of 38 replications of RTCM analyses. The natural logarithms of the fluorescence intensity values (corresponding to NF-κB activation) are depicted. The median value of GFP signals obtained with the empty-vector control (pcDNA) for each RTCM replication was set to 1 (black). Signals were sorted according to decreasing average fc. The color scheme from red to black to green corresponds to the fluorescence intensity values from high [ln(fc) = 3] to low [ln(fc) = −1.18]. The numbers in the bottom line (1 to 38) indicate the different replications. Replications with the same underlining were carried out on one slide. Three different preparations of spotted slides (screen series A to C) were used. (C) GFP signals corresponding to NF-κB activity were determined for all 38 replications by using a laser scanner (473 nm) and analyzed with the AIDA software package. Median values of relative fluorescence intensities (Rel. FI) compared to that of the empty-vector control pcDNA (set to 1; shaded gray) were calculated and are arranged in decreasing order (except for constitutive GFP signals and combinations of HHV-8 genes with the LMP-1 gene). Values obtained with certain genes which are of relevance in this report are indicated. Genes and gene combinations which resulted in more-than-twofold (fc ≥ 2.0) and highly significant (P ≤ 0.001) activation of NF-κB are given in the inset.