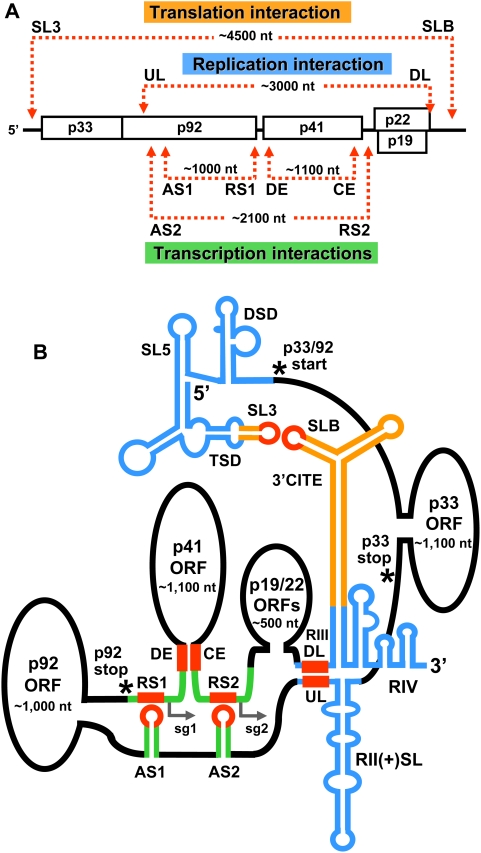
Figure 9. Linear and higher-order structural models for the functional long-distance RNA–RNA interactions that occur in the TBSV genome.
(A) Linear representation of the TBSV genome showing RNA–based interactions involved in translation, replication, and sg mRNA transcription. (B) Higher-order structural model for long-range RNA–based interactions in the TBSV genome. The sequences that are directly involved in forming long-range base pairing interactions are shown in red, while associated sequences and/or structures that are involved in translation, replication, and sg mRNA transcription are color coded as orange, blue, and green, respectively. Relevant structures are labeled (TSD, T-shaped domain; DSD, downstream domain; AS, activator sequence; RS, receptor sequence; CE, core element; DE, distal element). The SL3–SLB interaction is required for translation of p33/92 from the genome. The AS1–RS1 interaction is required for sg mRNA1 transcription, while both the AS2–RS2 and DE–CE interactions are required for sg mRNA2 transcription. The TSD, SL5, and DSD are located 5′-proximally in the 5′UTR of the genome and are involved in mediating genome plus-strand RNA synthesis [65]. Large intervening sections of sequence, which are predicted by mfold to form domains (see Figure S1), are shown as ovals and roughly correspond to the ORFs for p33, p92, p41, and p19/22 (not to scale). The start codon for p33/92, as well as the termination codons for these two proteins, are labeled and denoted by asterisks. Initiation sites of the two sg mRNAs are indicated by small arrows. See text for additional details.