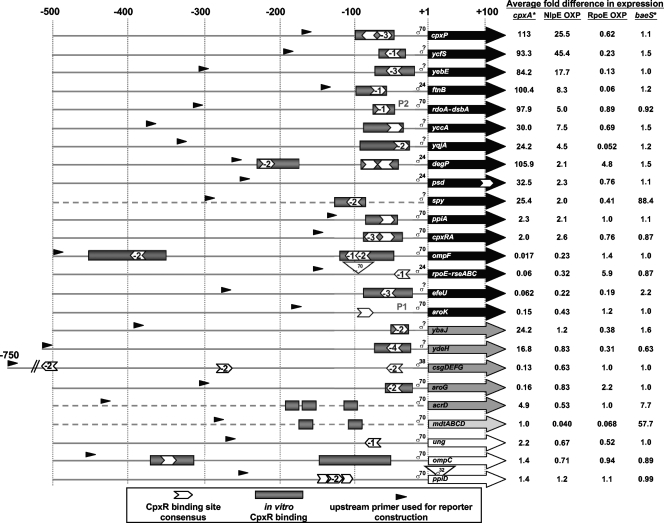
FIG. 1.
Evidence for Cpx regulation of the proposed Cpx regulon of E. coli MC4100. Numbers at the top indicate relative distance from the transcription start point for RNAP containing the sigma factor indicated next to the +1 site. Dashed lines indicate genes for which the transcription start site is not known. In these cases, +1 indicates the translational start site. Arrowheads indicate the position and orientation of the consensus CpxR binding site 5′ GTAAAN5GTAAA 3′. The numbers within the arrowheads indicate the number of nucleotides that differ from the consensus CpxR binding site. Shaded rectangles show the locations of demonstrated CpxR binding sites. Large arrows represent transcripts for the indicated genes. Black arrows represent genes shown to be significantly Cpx regulated under both strong (cpxA24 allele) and mild (NlpE overexpression) Cpx-activating conditions; dark gray arrows show genes that show Cpx regulation only by strong Cpx-inducing cues; light gray arrows indicate genes that show Cpx regulation only by mild Cpx induction; and white arrows show genes that are weakly or not Cpx regulated (as determined in this study). Small, black arrowheads indicate the location of the upstream primer used to construct the lux reporter for that gene. Numbers on the right indicate the average differences in expression in the various indicated backgrounds relative to the wild-type or respective vector controls. The average differences shown are based on the average differences observed for at least two different experiments assaying three replicates each time. Various backgrounds tested are cpxA* (cpxA24 allele), NlpE OXP (NlpE overexpression), RpoE OXP (σE overexpression), and baeS* (baeS1 allele).