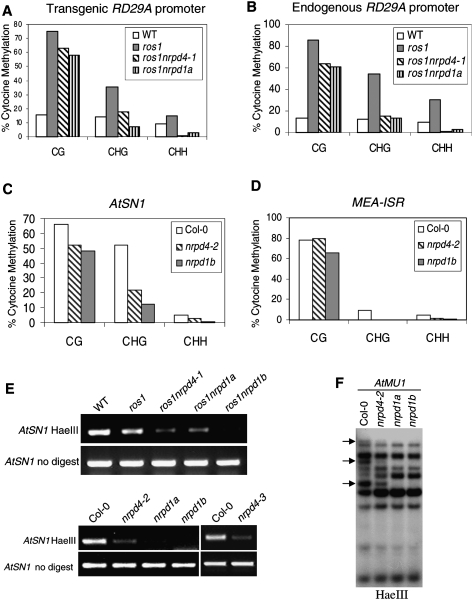
Figure 3.
Effect of nrpd4 mutations on DNA methylation. The percentage of cytosine methylation at transgene (A) and endogenous (B) RD29A promoters, and at AtSN1 (C) and MEA-ISR (D) were determined by bisulfite sequencing. The ros1nrpd1a or nrpd1b mutants were used as the mutant controls. (H) A, T, or C. The percentage of cytosine methylation on CG, CHG, and CHH sites is shown. (E) PCR assay of the effect of the nrdp4-1 mutation on DNA methylation of AtSN1. Amplification of AtSN1 was performed after the genomic DNA was digested with the methylation-sensitive restriction enzyme HaeIII. The undigested genomic DNA was amplified as a control. (F) Effect of the nrpd4 mutation on DNA methylation of AtMU1. Genomic DNA from Col-0, nrpd4-2, nrpd1a, and nrpd1b was digested with the methylation-sensitive restriction enzyme HaeIII, followed by Southern hybridization.