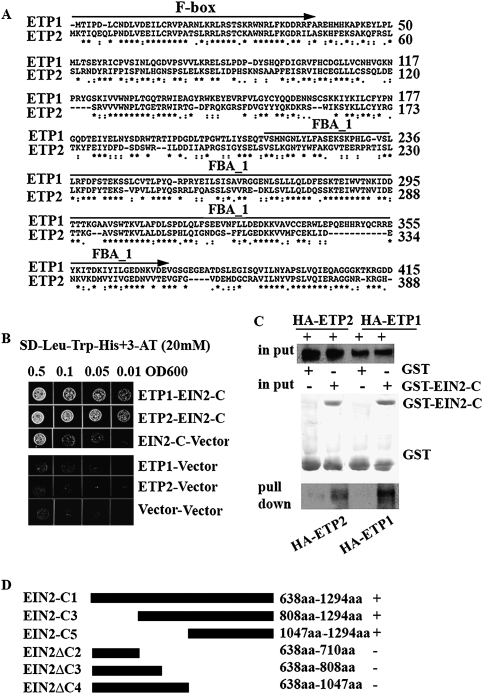
Figure 2.
Two novel F-box proteins, ETP1 and ETP2, interact with EIN2-CEND (EIN2-C). (A) Alignment of ETP1 and ETP2 amino acid sequences generated with the ClustalW program. The positions of amino acid residues are indicated with numbers; asterisks and dots indicate identical and conserved amino acids, respectively. The putative F-box motif and the FBA_1 (F-box protein-associated) domain are indicated by arrow above the sequences (B) EIN2-CEND interacts with ETP1 and ETP2 in yeast. Growth on selective plates lacking adenine, histidine, tryptophan with 20 mM 3-AT (−Leu, −Trp, −His, +3AT) and on control plates lacking only tryptophan and leucine (−Trp, −Leu) is shown. (C) EIN2 interacts with ETP1 and ETP2 in vitro. GST-EIN2-CEND (GST-EIN2-C) fusion protein and GST alone protein were purified from E. coli. These proteins, as well as GST alone, were assayed to pull-down with in vitro translated and HA-tagged ETP1 and ETP2 proteins. The same quantities of the GST fusion proteins (bottom panel) and the same amount of HA-tagged ETP1 or ETP2 (middle panel) were used as inputs. (Top panel) The HA-tagged ETP1 and ETP2 were detected by anti-HA antibody. A plus sign (+) indicates the addition of protein; a minus sign (−) indicates the protein was not added. (D) EIN2 highly conserved CEND (EIN2-C5) is sufficient to interact with ETP1 and ETP2 in yeast. The diagrams indicate different deletions of EIN2 CEND. A plus sign (+) indicates interaction; a minus sign (−) indicates no interaction. (WB) Western blot.