Abstract
Mutations in the genes PTEN-induced putative kinase 1 (PINK1), PARKIN, and DJ-1 cause autosomal recessive forms of Parkinson disease (PD), and the Pink1/Parkin pathway regulates mitochondrial integrity and function. An important question is whether the proteins encoded by these genes function to regulate activities of other cellular compartments. A study in mice, reported by Xiong et al. in this issue of the JCI, demonstrates that Pink1, Parkin, and DJ-1 can form a complex in the cytoplasm, with Pink1 and DJ-1 promoting the E3 ubiquitin ligase activity of Parkin to degrade substrates via the proteasome (see the related article beginning on page 650). This protein complex in the cytosol may or may not be related to the role of these proteins in regulating mitochondrial function or oxidative stress in vivo.
Parkinson disease (PD) is the second most common neurodegenerative disorder, and mutations in the genes PTEN-induced putative kinase 1 (PINK1, also known as Parkinson disease 6 [PARK6]), PARKIN (also known as PARK2), and DJ-1 (also known as PARK7) cause autosomal recessive forms of PD/parkinsonism. PINK1 encodes a protein with a mitochondrial targeting sequence and a putative serine/threonine kinase domain, and PINK1 is predominantly localized to mitochondria (1). The Parkin protein contains two RING finger motifs, has E3 ubiquitin ligase activity in vitro, and is largely localized to the cytosol (1). The endogenous DJ-1 protein is found in mitochondria and cytosol, but the function of DJ-1 is not entirely clear (1). Studies on the functions of these genes may provide important insights into PD pathogenesis.
Prior studies on protein degradation
Most PD patients have intraneuronal inclusions in the form of ubiquitin-positive Lewy bodies and Lewy neurites. Given the presence of Parkin in Lewy bodies and the putative role of Parkin as an E3 ligase, much of the initial work on Parkin was focused on its potential role in regulating protein degradation via the ubiquitin-proteasome system (UPS). Ubiquitination is accomplished by covalently linking the ubiquitin polypeptide (Ub) to a lysine residue in a specific protein substrate and requires the sequential action of an E1 activating enzyme, an E2 conjugating enzyme, and an E3 ligase (2). E3 ubiquitin ligases can mediate monoubiquitination (the addition of a single Ub to the substrate protein), multiubiquitination (the addition of multiple single Ubs to different lysine residues in a target protein), and polyubiquitination (in which chains of four or more Ubs are formed by the linkage of Ub molecules to lysine residues in other Ub molecules) (2). These linkages are most often to lysine 48 (K48) or lysine 63 (K63) of the Ub polypeptide. Proteins to be degraded by the proteasome are largely K48 polyubiquitinated. In contrast, K63 polyubiquitination, as well as monoubiquitination and multiubiquitination, primarily function in non-degradative processes including signal transduction, transcriptional regulation, protein localization, and membrane trafficking (2).
Previous studies suggested that Parkin could function as an E3 ligase for proteasome-mediated protein degradation (3). A handful of substrates of Parkin, including Parkin itself and an α-synuclein–interacting protein (Synphilin-1) (4), have been identified in vitro. If Parkin were indeed important in the degradative pathway in vivo, one would expect that the levels of its substrates should increase in Parkin-knockout mice. Unexpectedly, however, most of the substrates studied, including Synphilin-1, did not accumulate in Parkin-null mice (5). In addition, several studies have revealed that Parkin preferentially catalyzes monoubiquitination and K63-linked polyubiquitination of substrates including Synphilin-1 (6–8). These latter observations may offer potential explanations for the lack of substrate accumulation in vivo by implicating Parkin in a non-degradative, proteasome-independent process. Studies in Drosophila and more recently in mammals have provided important insights into Parkin function, although whether Parkin possesses degradative or non-degradative functions remains to be determined.
Central role of mitochondrial function in PD pathogenesis
Flies lacking Parkin function show striking defects in mitochondrial morphology that are highly similar, if not identical, to those observed in Pink1 mutants (9–11). Genetic epistasis experiments have demonstrated that Parkin and Pink1 act in a common genetic pathway, with Pink1 positively regulating Parkin (9, 10). This Pink1/Parkin pathway controls mitochondrial integrity at least in part via promotion of mitochondrial fission and/or inhibition of mitochondrial fusion (12–14). Consistent with these findings in Drosophila, patients with PINK1 or PARKIN mutations have indistinguishable clinical features and also show mitochondrial defects (1, 15). Recent studies also suggest that Pink1 and Parkin regulate mitochondrial functions in mammals (16–19). These findings underscore the central importance of the Pink1/Parkin pathway in regulating mitochondrial integrity and function.
Parkin is localized largely in the cytosol, though it can be found within mitochondria or associated with the outer mitochondrial membrane in certain contexts (20–22). Meanwhile, Pink1 has been found within mitochondria in cells and in vivo (9, 23, 24). The mechanism by which Parkin interacts with Pink1 and acts on mitochondria has remained a mystery. Interestingly, a recent study on the topology of Pink1 suggests that Pink1 may be anchored in the mitochondrial outer membrane, with its kinase domain facing toward the cytoplasm (25), providing one possible route for the Pink1/Parkin interaction.
Revisiting protein degradation in the cytosol
In this issue of the JCI, Xiong and colleagues report a possible mechanism by which Parkin, Pink1, and DJ-1 might function outside of mitochondria in mice (26). Previous work had suggested that Pink1 binds to DJ-1 (27), DJ-1 binds to Parkin (28), and Pink1 binds to Parkin (1, 29). Here, Xiong et al. extend these observations by showing that the 3 proteins co-fractionate in gel filtration assays and co-immunoprecipitation in vivo. These interactions were observed largely in the cytosol, rather than in the mitochondria. Moreover, when Pink1 or DJ-1 are overexpressed or provided directly in vitro, ubiquitination and degradation of Parkin substrates (Synphilin-1 and Parkin itself) via the UPS pathway were enhanced, while levels of ubiquitinated Parkin decreased in Pink1-mutant brain slice culture. The authors also show that pathogenic mutants of Pink1 and Parkin impaired the activity of the Parkin/Pink1/DJ-1 (PPD) complex, providing a potential mechanistic basis for how mutations in these proteins lead to PD pathogenesis. Based on these observations, the authors propose that Pink1, Parkin, and DJ-1 form a novel, cytoplasmically localized E3 ligase complex (the PPD complex) in which Pink1 and DJ-1 modulate Parkin-dependent ubiquitination and subsequent degradation of substrates. The authors also suggest that the PPD complex promotes the degradation of unfolded proteins. This conclusion is based largely on the observation that heat shock stress to cells in culture (which is presumed, though not shown, to cause misfolding of Parkin) resulted in increased ubiquitination and accumulation of Parkin, an effect that was suppressed when Pink1 was overexpressed. These results are intriguing, but heat shock is likely to alter the folding of many proteins and to affect the activity of myriad signaling pathways. Thus it is unclear whether the observed effects reflect direct interactions between Pink1, Parkin, and unfolded substrates. It will be particularly interesting to follow the fate of specific proteins known to be misfolded, when added to otherwise unperturbed cellular systems in which levels and activities of Pink1 and Parkin have been altered.
Possible connections between mitochondrial dysfunction and protein degradation?
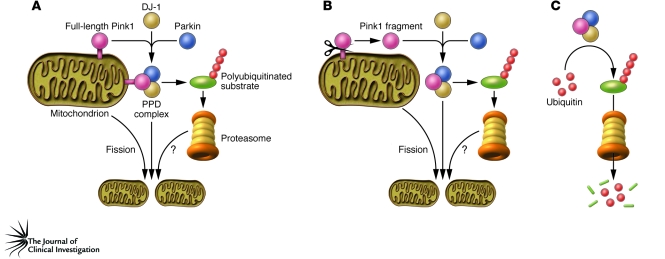
How might the function of the PPD complex be related to the established roles of Pink1 and Parkin in regulating mitochondrial integrity and function? It is possible that the roles of Pink1/Parkin in regulating mitochondrial function are related to the potential degradative functions of the PPD complex (Figure 1, A and B). DJ-1 can be recruited to the mitochondrial outer membrane following oxidative stress (30). Upon mitochondrial uncoupling, overexpressed Parkin can also be selectively recruited onto impaired mitochondria, promoting their removal through autophagy (31). If the kinase domain of Pink1 does indeed face the cytoplasm (25), Parkin and DJ-1 might be recruited to the mitochondrial surface under certain conditions and may thereby interact with Pink1 at the mitochondrial outer membrane (Figure 1A). Perhaps this interaction facilitates the ubiquitin ligase activity of Parkin, thereby promoting the turnover of molecules that regulate mitochondrial dynamics and mitophagy. A complication with this model, however, is that Xiong and colleagues report that while full-length Pink1 localized to mitochondria, a processed fragment of Pink1 was found predominantly in the cytoplasm, with this latter fragment being the version of Pink1 that predominantly interacts with Parkin (26). Similar cytosolic Pink1 fragments have been reported in earlier studies (32), suggesting a model in which Pink1 is cleaved and released from mitochondria under certain stress conditions. Cleaved forms of Pink1 may thus be able to interact with Parkin in the cytosol and thereby promote degradative ubiquitination or other cellular processes (Figure 1B). Pink1-dependent activities in the cytoplasm may still serve to regulate mitochondrial function, for example by relaying information about mitochondrial status to other cellular compartments. The fact that a PD-causing mutation in the PINK1 kinase domain, G309D, compromises both the PPD complex function and mitochondrial integrity in Drosophila muscle and male germline (33) is consistent with the hypothesis that the PPD complex may regulate mitochondrial function. Alternatively, cytosolic Pink1 may carry out mitochondria-independent functions, perhaps also in complex with Parkin and DJ-1 (Figure 1C). It is important to note that loss of DJ-1 results in phenotypes that are distinct from the mitochondrial defects associated with loss of Pink1/Parkin in Drosophila (9, 10, 34). If DJ-1 is an obligatory component of this complex, these in vivo results would suggest that the PPD complex is not essential for regulation of mitochondrial integrity. Future experiments are needed to clarify the role of DJ-1 function.
Figure 1. Three models for the role of the PPD complex.
In this issue of the JCI, Xiong et al. report that Pink1, Parkin, and DJ-1 bind to each other and form a PPD E3 ligase complex in which Pink1 and DJ-1 modulate Parkin-dependent ubiquitination and subsequent degradation of substrates via the proteasome (26). Previous work suggests that the Pink1/Parkin pathway regulates mitochondrial integrity and promotes mitochondrial fission in Drosophila (9–14). (A) Parkin and DJ-1 may be recruited to the mitochondrial outer membrane during stress (30, 31) and interact with Pink1. These interactions may facilitate the ligase activity of Parkin, thereby facilitating the turnover of molecules that regulate mitochondrial dynamics and mitophagy. The PPD complex may have other roles in the cytosol that result in degradative ubiquitination (26) and/or relay information from mitochondria to other cellular compartments. (B) Alternatively, Pink1 may be released from mitochondria after cleavage to interact with DJ-1 and Parkin in the cytosol. A and B differ in the site of action of the PPD complex and the cleavage status of Pink1. The complex forms on the mitochondrial outer membrane potentially containing full-length Pink1 in A, and in the cytosol with cleaved Pink1 in B. The question marks indicate speculative connections. Note that lack of DJ-1 function results in phenotypes that are distinct from the mitochondrial phenotypes observed in null mutants of Pink1 or Parkin in Drosophila (9, 10, 12, 13, 34). Thus, although the PPD complex is illustrated here as regulating mitochondrial fission, the role of DJ-1 in vivo remains to be clarified. (C) It is also possible that the action occurs in the cytosol and is independent of the function of Pink1/Parkin in regulating mitochondrial integrity and function.
Summary
In summary, while the current study by Xiong et al. (26) rekindles enthusiasm for exploring the roles of Parkin in mediating protein degradation via the UPS, it remains to be shown whether Parkin, in complex with Pink1 and DJ-1, carries out protein degradation in vivo. It will be particularly important to reconcile the observation by the authors that the PPD complex degrades Synphilin-1, with the in vivo observations demonstrating that Synphilin-1 levels do not accumulate in knockout mice lacking Parkin (5). Perhaps the PPD complex only functions to regulate Synphilin-1 levels when cells are stressed, but not under the conditions assayed in the Parkin-knockout mice. The Xiong et al. study offers an entry point for explorations of the role of Pink1, Parkin, and DJ-1 in the cytoplasm. The priority now becomes identifying in vivo contexts in which this complex functions.
Acknowledgments
We apologize to those whose work we were unable to cite because of space constraints. We thank Mark Dodson and Bruce Hay for helpful comments on the manuscript. This work is supported by funds from the McKnight Foundation, Alfred P. Sloan Foundation, Klingenstein Fellowship, American Parkinson Disease Association, and NIH (R01, P01, and K02 grants) to M. Guo.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: PARK2, Parkinson disease 2; PD, Parkinson disease; PINK1, PTEN-induced putative kinase 1; PPD, Parkin/Pink1/DJ-1 (complex); Ub, ubiquitin polypeptide; UPS, ubiquitin-proteasome system.
Citation for this article: J. Clin. Invest. 119:442–444 (2009). doi:10.1172/JCI38619.
See the related article beginning on page 650.
References
- 1.Dodson M.W., Guo M. Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson’s disease. Curr. Opin. Neurobiol. 2007;17:331–337. doi: 10.1016/j.conb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Mukhopadhyay D., Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–215. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 3.Kahle P.J., Haass C. How does parkin ligate ubiquitin to Parkinson’s disease? EMBO Rep. 2004;5:681–685. doi: 10.1038/sj.embor.7400188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung K.K., et al. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat. Med. 2001;7:1144–1150. doi: 10.1038/nm1001-1144. [DOI] [PubMed] [Google Scholar]
- 5.Ko H.S., et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J. Neurosci. 2005;25:7968–7978. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim K.L., et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J. Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuda N., et al. Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J. Biol. Chem. 2006;281:3204–3209. doi: 10.1074/jbc.M510393200. [DOI] [PubMed] [Google Scholar]
- 8.Olzmann J.A., et al. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark I.E., et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 10.Park J., et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y., et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng H., Dodson M.W., Huang H., Guo M. The Parkinson’s disease genes pink1 and parkin promotes mitochondrial fission and/or inhibits mitochondrial fusion. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poole A.C., et al. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y., et al. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibanez P., et al. Mutational analysis of the PINK1 gene in early-onset parkinsonism in Europe and North Africa. Brain. 2006;129:686–694. doi: 10.1093/brain/awl005. [DOI] [PubMed] [Google Scholar]
- 16.Exner N., et al. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J. Neurosci. 2007;27:12413–12418. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gautier C.A., Kitada T., Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piccoli C., et al. Mitochondrial respiratory dysfunction in familiar parkinsonism associated with PINK1 mutation. Neurochem. Res. 2008;33:2565–2574. doi: 10.1007/s11064-008-9729-2. [DOI] [PubMed] [Google Scholar]
- 19.Wood-Kaczmar A., et al. PINK1 is necessary for long term survival and mitochondrial function in human dopaminergic neurons. PLoS ONE. 2008;3:e2455. doi: 10.1371/journal.pone.0002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimura H., et al. Immunohistochemical and subcellular localization of Parkin protein: absence of protein in autosomal recessive juvenile parkinsonism patients. Ann. Neurol. 1999;45:668–672. doi: 10.1002/1531-8249(199905)45:5<668::AID-ANA19>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 21.Darios F., et al. Parkin prevents mitochondrial swelling and cytochrome c release in mitochondria-dependent cell death. Hum. Mol. Genet. 2003;12:517–526. doi: 10.1093/hmg/ddg044. [DOI] [PubMed] [Google Scholar]
- 22.Kuroda Y., et al. Parkin enhances mitochondrial biogenesis in proliferating cells. Hum. Mol. Genet. 2006;15:883–895. doi: 10.1093/hmg/ddl006. [DOI] [PubMed] [Google Scholar]
- 23.Silvestri L., et al. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum. Mol. Genet. 2005;14:3477–3492. doi: 10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- 24.Gandhi S., et al. PINK1 protein in normal human brain and Parkinson’s disease. Brain. 2006;129:1720–1731. doi: 10.1093/brain/awl114. [DOI] [PubMed] [Google Scholar]
- 25.Zhou C., et al. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong H., et al. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J. Clin. Invest. . 2009;119:650–660. doi: 10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang B., et al. Association of PINK1 and DJ-1 confers digenic inheritance of early-onset Parkinson’s disease. Hum. Mol. Genet. 2006;15:1816–1825. doi: 10.1093/hmg/ddl104. [DOI] [PubMed] [Google Scholar]
- 28.Moore D.J., et al. Association of DJ-1 and parkin mediated by pathogenic DJ-1 mutations and oxidative stress. Hum. Mol. Genet. 2005;14:71–84. doi: 10.1093/hmg/ddi007. [DOI] [PubMed] [Google Scholar]
- 29.Kim Y., et al. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem. Biophys. Res. Commun. 2008;377:975–980. doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 30.Canet-Aviles R.M., et al. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beilina A., et al. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5703–5708. doi: 10.1073/pnas.0500617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yun J., et al. Loss-of-function analysis suggests that Omi/HtrA2 is not an essential component of the PINK1/PARKIN pathway in vivo. . J. Neurosci. 2008;28:14500–14510. doi: 10.1523/JNEUROSCI.5141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meulener M., et al. Drosophila DJ-1 mutants are selectively sensitive to environmental toxins associated with Parkinson’s disease. Curr. Biol. 2005;15:1572–1577. doi: 10.1016/j.cub.2005.07.064. [DOI] [PubMed] [Google Scholar]