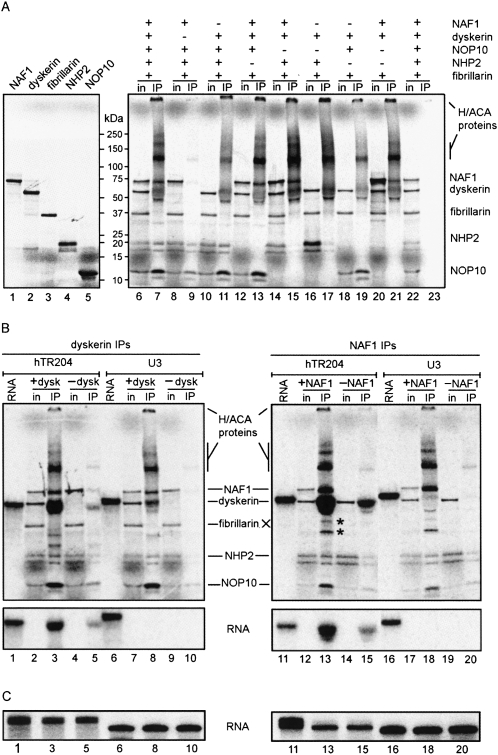
FIGURE 2.
The tetramer NAF1-dyskerin-NOP10-NHP2 assembles specifically with hTR204. (A) Individual proteins were translated in vitro in the presence of 35S-labeled methionine and analyzed by PAGE (left). For dyskerin IPs (right), 35S-labeled proteins were cotranslated; the presence or absence of each protein in the RRL mixture is indicated by (+) or (−), respectively. Input of translated proteins (in) and immunoprecipitated products (IP) were separated by PAGE. Molecular weight markers (kDa) are shown between the two panels. Control IP with beads alone is shown (lanes 22,23). Note that dyskerin recovered from IPs gives a fuzzy signal. High molecular weight multimers of H/ACA proteins are also observed. (B) Dyskerin IPs (left) and NAF1 IPs (right) of RRL mixtures containing 35S-labeled proteins and 32P-labeled hTR204 (lanes 1-5,11-15) or U3 (lanes 6-10,16-20) RNAs were analyzed by PAGE. Because RNAs co-migrate with dyskerin in these protein gels, input lanes (in) did not contain the 32P-labeled RNAs to better discern the 35S-labeled proteins. IPs with RRL mixtures lacking either dyskerin (-dysk) or NAF1 (-NAF1) were used as negative controls. Labeled hTR204 (lanes 1,11) and U3 (lanes 6,16) RNAs not incubated in RRL are shown. Bands marked by an asterisk (*) correspond to degradation products of dyskerin/NAF1, not to fibrillarin, which was omitted form NAF1 IPs (right). (Bottom) The 32P signal from the protein gels presented in the top panels; they were obtained by blocking the 35S signal with a transparency. (C) Labeled RNAs from input (lanes 1,6,11,16) and IP supernatants (lanes 3,5,8,10,13,15,18,20) were analyzed separately on 8% denaturing polyacrylamide gels. Lanes are numbered in accordance with the corresponding IPs shown in (B).