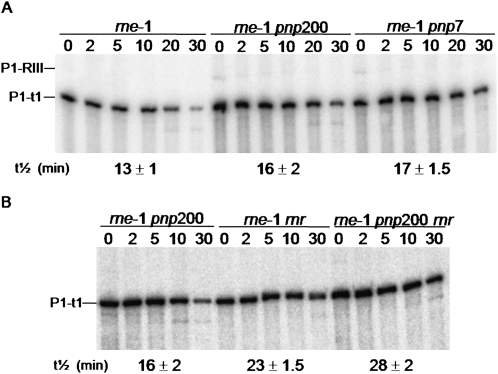
FIGURE 1.
Effect of RNase R and PNPase on the degradation of the rpsO mRNA. (A) Analysis of the rpsO mRNA stability upon PNPase depletion. Total RNA from rne-1, rne-1 pnp200, and rne-1 pnp7 was extracted from exponential phase cultures grown at 30°C and incubated for 30 min at the nonpermissive temperature of 44°C, before inhibition of transcription. Two alleles coding for PNPase were tested: the pnp200 encodes the thermolabile PNPasets (Yancey and Kushner 1990) while the pnp7 is a nonsense mutant (Mohanty and Kushner 2003). The P1-t1 mRNA corresponds to the monocistronic rpsO transcript while the P1-RIII mRNA results from the processing of the rpsO-pnp bicistronic transcript (Hajnsdorf et al. 1994). An antisense riboprobe against the full-length rpsO mRNA was used. Half-lives are expressed in minutes. (B) rpsO mRNA decay in the absence of PNPase and/or RNase R. Cultures of rne-1 pnp200, rne-1 rnr, and rne-1 pnp200 rnr strains were grown at 30°C to exponential phase and shifted to 44°C; after 30 min of heat incubation, rifampicin was added and culture samples were withdrawn at times indicated. All strains were studied under the same conditions. Five micrograms of total RNA were analyzed on a 6% polyacrylamide Northern blot. Half-lives are depicted at the bottom the corresponding strain and are expressed in minutes.