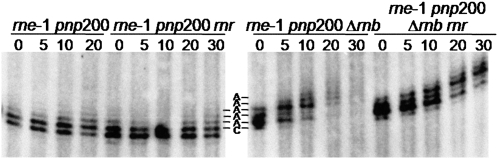
FIGURE 3.
RNase R removes the adenosine residues added by PAP I at the 3′ end of the rpsO transcripts upon RNase II inactivation. All strains carry the pnp200 allele and were exposed to thermal inactivation for 30 min prior to addition of rifampicin. Sample time points were withdrawn at times indicated. Ten micrograms of RNA extracted from rne-1 pnp200, rne-1 pnp200 rnr, rne-1 pnp200 Δrnb, and rne-1 pnp200 Δrnb rnr strains were mixed with the chimeric DNA/RNA CrpsO oligonucleotide, which binds ∼50 nt upstream of the terminal transcription residues of the rpsO transcripts and targets them to RNase H cleavage. The mixture was then incubated with RNase H. 3′ terminal fragment of rpsO transcripts were resolved on a 10% polyacrylamide gel and identified by hybridization with a riboprobe specific to the 3′ end of the rpsO mRNA. A run-off transcript matching the nonadenylated full-length rpsO was treated in the same conditions (not shown) (Marujo et al. 2000). RNA fragments that end at the transcriptional termination C residue were identified and are indicated (Left panel/right panel: corresponds, respectively, to the second and first band observed counting from the bottom). Longer RNA fragments correspond to RNAs harboring additional adenosine residues, indicated by the letter A.