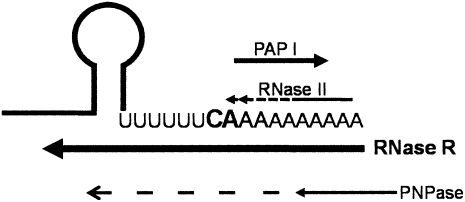
FIGURE 7.
Model of the poly(A)-dependent metabolism of the rpsO mRNA. The 3′ end of the rpsO transcript has a transcriptional termination hairpin followed by a short linear sequence. The major transcriptional termination nucleotides (CA) are in bold. RNase II removes very efficiently the nucleotides added by PAP I downstream from this sequence but does not proceed through the mRNA body (Marujo et al. 2000; Folichon et al. 2005). When RNase II is inactive, longer poly(A) tails arise promoting the degradation of the rpsO mRNA. Polyadenylation extends the short 3′ linear region, providing a toehold for exonucleases. RNase R and PNPase may degrade this transcript, although with different specificities. RNase R requires a less extensive 3′ end linear region to efficiently bind to RNA than PNPase. Addition of two nucleotides after the (U)6CA fragment are enough to initiate RNase R degradation. This becomes advantageous in the competition with PNPase for the access to the 3′ end of the rpsO mRNA. The growing poly(A) tails favor RNase R degradation. In the presence of poly(A) tails RNase R can easily overcome the RNA secondary structure and completely degrade the rpsO mRNA. RNase R is therefore established as a major poly(A)-dependent exonuclease involved in the degradation of the rpsO mRNA. Polyadenylated RNAs can be attacked by RNase II and RNase R preventing the appearance of longer poly(A) tails that can act as suitable substrate to PNPase. Only if RNase II and RNase R are inactive, longer poly(A) tails might arise that can either be shortened (arrow) or possibly promote RNA degradation by PNPase (dotted arrow).