Fig 1.
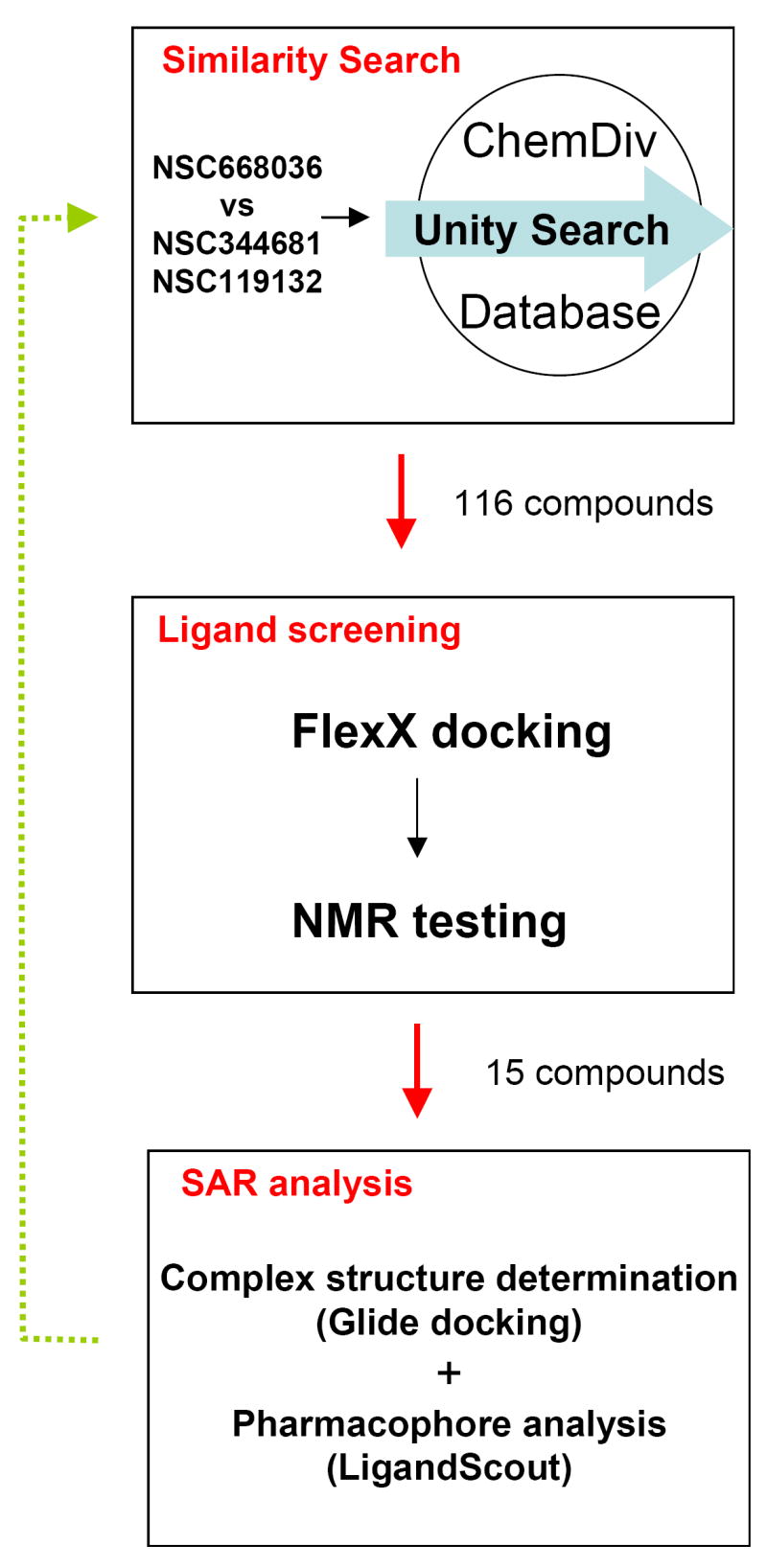
The workflow of optimizing PDZ domain inhibitors by exploring chemical space. Queries designed based on a PDZ-NSC668036 complex was used to search ChemDiv database for PDZ domain inhibitors by using Unity 2D/2D similarity searches. Returned 116 hits were docked using FlexX and 15 compounds were selected for NMR testing and their docking conformations were refined by Glide (Schrödinger Inc.). Combining NMR testing, Glide optimization of docking poses and LigandScout (Inte:Ligand, Austria) pharmacophore analysis, we derived SAR for the 15 new PDZ domain inhibitors.
