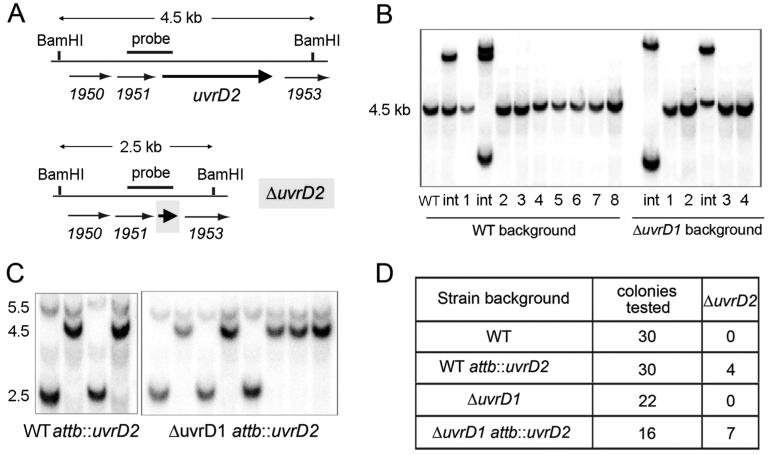
Figure 10.
Evidence that UvrD2 is essential in M. smegmatis. (A) Schematic maps of the wild-type M. smegmatis genomic uvrD2 locus (MSmeg_1952), indicated by the thick arrow, with flanking ORFs depicted as thin arrows. BamHI restriction sites and the genomic segment used as a radiolabeled probe for genotyping are indicated. A map of the ΔuvrD2 locus generated by targeted insertion of an in-frame deletion of the uvrD2 ORF is shown below the wild-type locus map. (B) BamHI digests of genomic DNA from wild-type M. smegmatis and the ΔuvrD1 mutant, hygR sucS merodiploids generated by integration of the ΔuvrD2 cassette at the uvrD2 locus (int), and individual hygS sucR segregants derived from the merodiploids were resolved by agarose gel electrophoresis, transferred to a membrane, and hybridized to a 32P-labeled DNA probe generated by amplification of the segment depicted in panel A. All of the segregants retained the 4.5-bp BamHI fragment corresponding to the wild-type uvrD2 locus. (C) BamHI digests of genomic DNA from individual hygS sucR segregants derived from the merodiploids of attB::uvrD2 and ΔuvrD1 attB::uvrD2 strains of M. smegmatis containing the ΔuvrD2 cassette integrated at the uvrD2 locus were genotyped by Southern blotting as described in panel B. Segregants containing only the 2.5 kb ΔuvrD2 locus were recovered when a second uvrD2 allele was preintegrated at the genomic attB site. (D) Summary of the outcomes of attempts to delete uvrD2 in M. smegmatis strains that do or do not have an extra copy of uvrD2 integrated into the chromosomal attB locus.