Abstract
Entamoeba histolytica is an important human pathogen and a leading parasitic cause of death globally. The parasite life cycle alternates between the trophozoite form, which is motile and causes invasive disease and the cyst stage, which is environmentally resistant and transmits infection. Understanding the triggers that initiate stage conversion is an important yet understudied area of investigation. Recent progress in dissecting the transcriptional networks that regulate E. histolytica development is outlined in this review.
Keywords: Microarray, Gene expression, Cyst, Trophozoite, Encystation
1. Introduction
Entamoeba histolytica is a pathogenic amoeba that is widespread throughout the world but is especially common in the developing world, where many people do not have access to clean water supplies. The parasite causes invasive disease in over 50 million people and an estimated 100,000 deaths per year, making it a leading cause of parasitic death in humans (WHO, 1997). In Dhaka, Bangladesh, where diarrheal diseases are the leading cause of death in children younger than 6 years of age, ~ 50% of children have serological evidence of exposure to E. histolytica by age 5 years (Haque et al., 1999). Infection with E. histolytica can lead to asymptomatic colonization, amebic colitis or extra-intestinal disseminated disease. The most common extra-intestinal disease attributed to E. histolytica is liver abscess but infection may spread to other organs, particularly the lung and brain (Haque et al., 2003). Several virulence factors have been identified that are important for parasite pathogenesis, however the majority of molecular mechanisms that the parasite utilizes as it establishes invasive disease remain unclear (Stanley, 2003).
The infectious cycle of E. histolytica begins with the ingestion of the cyst, a non-dividing, quadrinucleate form that is able to survive in the environment due to a protective, chitin-containing cell wall (Haque et al., 2003). After ingestion, the cyst undergoes excystation in the small intestine to produce the proliferative trophozoite form. Trophozoites colonize the colon, adhering to the mucus layer by means of a lectin, which binds colonic mucins. Disease results when this mucus layer is penetrated and the trophozoites attack the intestinal epithelial cells. Due to unknown factors, some trophozoites encyst, allowing them to be excreted in the stool and to go on to infect new hosts. Since only the cyst form can be transmitted, blocking encystation would prevent new infections, making the encystation program an attractive target for the development of new drugs or a vaccine.
Early studies of E. histolytica utilized xenic cultures in which the protozoa were grown in a complex medium with polymicrobial bacterial flora. Under these conditions, spontaneous encystation was observed (Jensen, 1983). However, establishment of axenic culture methods (or methods in which the xenic flora were less complex, largely aerobic or monomicrobial) greatly facilitated Entamoeba research, but led to the loss of the ability to differentiate into cysts (Eichinger, 1997). Long-term axenic growth in tissue culture has undoubtedly selected for strains that are able to adapt to those conditions, but has likely selected against the ability to differentiate into cysts. Although there have been published reports of formation of chitin containing cyst-like structures in response to certain stimuli, such as metal ions and serum (Chayen et al., 1988; Gonzalez-Salazar et al., 2000), these structures are likely not fully differentiated cysts, as they have a defective cell wall and are primarily mononucleate. These are likely pre-cysts that could not differentiate fully. To date, no method for high efficiency in vitro encystation with production of mature cysts is available.
Due to lack of an in vitro encystation system in E. histolytica, most research on encystation of Entamoeba has been performed using the related reptilian parasite, Entamoeba invadens. In E. invadens, encystation in vitro can be triggered using either osmotic shock (Bailey and Rengypian, 1980), or removal of glucose or other carbon sources (Vazquezdelara-Cisneros and Arroyo-Begovich, 1984; Byers et al., 2005). However, achieving high levels of cyst formation requires the addition of either 5% serum or the glycoprotein mucin, which acts as a ligand for the Entamoeba membrane lectin and stimulates the aggregation of trophozoites (Eichinger, 2001). This phenomenon is dose-specific as high concentrations of mucin can inhibit aggregation and encystation. This requirement for mucin can be circumvented by treatment with β-adrenergic receptor agonists, suggesting a function for this pathway in the regulation of encystation (Coppi et al., 2002). Recent work has indicated a link between colonic short chain fatty acids (SCFAs) and inhibition of parasite encystation (Byers et al., 2005). Since glucose is not present in the enteric environment, but SCFAs are produced by colonic microbial flora, it is interesting to speculate that the carbon source that the parasites are exposed to has effects on encystation potential. Roles for cell cycle regulation and proteasome function in E. invadens encystation have also been identified (Gangopadhyay et al., 1997; Gonzalez et al., 1999). In addition to these discoveries of encystation stimuli, several proteins expressed in cysts or in encysting trophozoites have been identified, including chitin synthase (Das and Gillin, 1991), chitinase (Villagomez-Castro et al., 1992), a glycoprotein that is a major component of cyst cell walls (Frisardi et al., 2000), and a developmentally regulated gene (Sanchez et al., 1994). Furthermore, a number of drugs have been identified as inhibiting excystation in E. invadens including inhibitors of cysteine proteases, protein kinase C, phosphatidylinositol 3-kinase and calcium chelators (Makioka et al., 2002a, b, 2003, 2005).
2. Recent advances in characterizing amebic development
During the last few years, our understanding of the molecular basis of Entamoeba development has improved substantially. These advances, largely predicated using information and tools based on the genome sequencing projects of E. histolytica (Loftus et al., 2005) and E. invadens (Wang et al., 2003), are outlined below.
2.1. Molecular analysis of Entamoeba cysts
Progress in characterizing Entamoeba development has been made with an improved understanding of the components of the cyst wall (Das et al., 2006; Van Dellen et al., 2006a, 2006b), histone modification (Byers and Eichinger, 2008), cellular control of excystation (Makioka et al., 2006) and handling of cellular debris (Picazarri et al., 2008).
The Entamoeba cyst wall is largely composed of proteins with chitin-binding domains, such as chitinases, Jessie lectins and Jacob lectins. The Jacob lectins are differentially modified by a number of post-translational modifications including proteolysis by cysteine proteases and glycosylation by O-phosphodiester-linked glycans (Van Dellen et al., 2006b). Overall, structural analysis of the cyst wall allows for the identification of potential novel diagnostic or therapeutic targets. SCFAs, which are released by bacteria inhabiting the human colon, have roles in Entamoeba development and enter the amebic trophozoites in a pH-dependent manner. Subsequent effects on acetylation of the amebic histone H4 N-terminal domain were demonstrated for a number of E. histolytica strains (Byers and Eichinger, 2008). Thus, the environmental milieu of the host colon and the bacterial by-products likely effect amebic gene expression and may have roles in regulating development in vivo. Furthermore, the requirement for galactose ligands during encystation was identified, indicating the complexity of signals and stimuli that affect parasite stage conversion (Turner and Eichinger, 2007). A report on the formation of chitin containing cyst-like structures in response to bacteria and histamine is similar to those referenced above in that the structures shown are likely not fully differentiated cysts (Barron-Gonzalez et al., 2008). As cells undergo development, they must have means of dealing with cellular debris. It has recently been demonstrated that the cellular mechanism of autophagy, a method of degrading damaged or unnecessary proteins and organelles, is functional in Entamoeba trophozoites and cysts (Picazarri et al., 2008). An Atg8 (autophagy related) conjugation system was identified in the genome sequence and an increase in Atg8-associated structures was noted in E. invadens trophozoites preceding the morphological and biochemical changes associated with early development, indicating that this mechanism may be a prerequisite for encystation.
2.2. Whole genome transcriptional profiling of E. histolytica cysts
Transcriptional profiling using microarray technology has revolutionized the way in which expression patterns are obtained. A number of different microarray approaches have been developed and utilized for studying important aspects of E. histolytica biology including strain differences (MacFarlane and Singh, 2006; Davis et al., 2007), host colonic and hepatic invasion (Gilchrist et al., 2006; Santi-Rocca et al., 2008), and stress response (Weber et al., 2006; Hackney et al., 2007). One approach that has been highly successful is the use of short oligonucleotides microarrays, based on Affymetrix technology (Gilchrist et al., 2006). Using this approach, Ehrenkaufer and colleagues developed the first whole-genome transcriptome of E. histolytica cysts (Ehrenkaufer et al., 2007b). As outlined above, recent clinical isolates of E. histolytica, when maintained in a complex diphasic media, retain their ability to generate cysts. Ehrenkaufer et al. (2007) used clinical samples isolated from patients which were maintained in culture for less than 8 weeks and were able to demonstrate the presence of cysts using calcofluor staining (calcofluor stains chitin in the cyst wall). This material was used for whole-genome expression profiling. Success was dependent on the ability to use extremely small amounts of RNA and utilize methods of signal amplification (Dumur et al., 2004), since the clinical isolates do not grow in large quantities.
By comparing the transcriptomes of two genetically distinct recent clinical isolates, which contain cysts, with expression data from a number of E. histolytica strains and conditions (including trophozoites of virulent and non-virulent strains and parasites harvested from a mouse model of amebic colitis), a subset of genes regulated uniquely in the samples that contain cysts were identified: genes whose expression increased in the clinical samples compared with controls were considered to be “cyst-specific” and genes whose expression decreased in the clinical samples compared with controls were considered to be “trophozoite-specific” (Ehrenkaufer et al., 2007b). Overall, 672 genes were cyst-specific and 767 genes were trophozoite-specific, indicating that ~ 15% of the E. histolytica genome is transcriptionally modulated during stage conversion. This extent of transcriptional modulation is similar to the extent of changes seen with developmental regulation in yeast (Chu et al., 1998).
Developmentally regulated genes identified in this manner include numerous cysteine proteases, of which both cyst- and trophozoite-specific examples were found, as well as several known virulence factors, which were all more highly expressed in trophozoites. In addition, numerous genes with possible signaling or regulatory roles were found to have developmentally regulated expression, including transmembrane kinases, calcium binding proteins and putative transcription factors. The proteins encoded by some of these genes may play a role in regulating stage conversion. The identification of cyst-specific genes will allow development of novel diagnostic and treatment approaches targeting cysts. Furthermore, the identification of the cyst transcriptome sets the stage for further characterization of the molecular basis of the encystation pathway.
2.3. Identification of stimuli that induce encystation
2.3.1. Complex culture medium
It has previously been described that complex medium (such as Robinson’s medium or LYSGM), which contain a complex bacterial flora, can support E. histolytica encystation (Balamuth, 1951; Chayen et al., 1988; Stechmann et al., 2008; Robinson, 1968) (http://homepages.lshtm.ac.uk/entamoeba/xenic.htm). The work of Ehrenkaufer and colleagues in developing the E. histolytica cyst transcriptome brought that information back to center stage. Additionally, they demonstrated that E. invadens spontaneously encysts with high efficiency in a complex medium, without the need for the osmotic shock or other stresses typically utilized to induce encystation (Ehrenkaufer et al., 2007b). Furthermore, the complex medium appears capable of maintaining long-term cultures of E. invadens; thus, the parasites appear to cycle between cysts and trophozoites, an observation that has not been recapitulated with E. invadens in other encystation media (G. Ehrenkaufer and D. Eichinger, unpublished data). For work with the recent clinical isolates of E. histolytica, Ehrenkaufer utilized Robinson’s medium (Robinson, 1968), but similar results with other E. histolytica clinical isolates have also been seen with LYSGM medium (C. G. Clark, personal communication). Thus, it appears that some components of the complex medium are sufficient to trigger developmental conversion in Entamoeba.
Because the complex media are able to support growth of Entamoeba cysts, we attempted to adapt E. histolytica laboratory isolates to Robinson’s medium. Unfortunately, all laboratory strains we tested (E. histolytica HM-1:IMSS, E. histolytica Rahman and E. histolytica 200:NIH), which have long been adapted to axenic growth, were not able to adapt to growth in Robinson’s medium. Regardless of the method of adaptation into Robinson’s medium (immediate or gradual adaptation), all laboratory strains died within 2–3 weeks of association with Robinson’s medium (Ehrenkaufer et al., 2007b; G. Ehrenkaufer and U. Singh, unpublished data). We also attempted to adapt parasites to xenic growth prior to growth in Robinson’s medium in order to get them acclimated to a phagocytic rather than pinocytic lifecycle, but met with no success. Parasite death under these conditions was not associated with the transition to a cyst, as determined by lack of development of a chitin-containing cyst wall and calcofluor staining. Similar observations were made with LYSGM medium with an inability to grow some axenic laboratory strains in this medium (C. G. Clark, personal communication).
2.3.2. Clinical strains maintained in complex xenic medium are most likely to be encystation competent
Initially we hypothesized that only recent clinical isolates would maintain encystation competence and that long-term in vitro culture would remove this stage conversion phenotype. However, we have maintained clinical isolates as xenic cultures with complex bacterial flora for more than 3 years without losing encystation competence (G. Ehrenkaufer, U. Singh and C. G. Clark, unpublished data). These findings are important in that they allow long-term studies on some aspects of ameba development with a given clinical strain. However, it appears that once an E. histolytica strain is axenized it may lose encystation competence. We obtained a clinical isolate when it was grown in complex medium and after it was axenized. Attempts to re-associate the axenic strain with bacterial flora and/or grow it in Robinson’s medium were not adequate to restore the ability to make cysts (G. Ehrenkaufer and U. Singh, unpublished data). We also compared the transcriptome of a recent axenic isolate in Robinson’s medium to a long-term axenic isolate in Robinson’s medium, and found that the more recently isolated strain did not have a molecular signature that was more reminiscent of cysts (G. Ehrenkaufer and U. Singh, unpublished data).
However, some E. histolytica isolates, even after having been axenized, are able to produce low numbers of cysts when returned to xenic growth in LYSGM medium (C. G. Clark, personal communication). The variables that determine the ability of an E. histolytica isolate to maintain encystation competence appear to include the parasite strain, bacterial flora, concentration of rice starch in the medium and the length of time a strain has been grown axenically. However, the results of attempts to induce encystation in xenic culture are highly variable, thus making it necessary to develop more robust techniques for in vitro encystation.
Axenization is a complicated process and undoubtedly represents a major bottleneck to parasite growth and survival. The parasites that survive this process are those that are able to adapt to growth under axenic conditions, and likely represent an extremely small minority of the starting culture (Diamond, 1961). It is not known whether parasites that survive the axenization process have rearrangements, deletions or point mutations of their chromosomes, which facilitate axenic growth but are detrimental to subsequent attempts at encystation. Another unlikely possibility is that the axenization bottleneck selects for very limited clones from a starting population, which were never encystation competent. With the advent of improved sequencing technologies, whole genome sequencing of clinical isolates before and after the axenization process can be undertaken to begin to answer these questions.
2.3.3. Contribution of bacterial flora
The presence of bacteria appears to be a requirement to obtain E. histolytica cysts in vitro, although this requirement does not apply to the in vitro encystation system for E. invadens (Bailey and Rengypian, 1980). The contribution of bacterial flora is not well understood. However, since E. histolytica encystation in the human host occurs in the presence of the colonic bacterial flora, a number of potential roles can be hypothesized including the contribution of SCFAs to cellular signaling, surface receptor interaction and nutrient acquisition as well as the influence of by-products of bacterial metabolism that are excreted in the culture medium. A bacterial flora, NRS, isolated from a monkey that was infected with a human isolate of E. histolytica (Dobell, 1928), was previously shown to be conducive for induction of encystation in vitro (Balamuth and Wieboldt, 1951). Unfortunately the components of that mixture are unknown and that flora has subsequently been lost (D. Eichinger, personal communication). When growing E. histolytica clinical isolates, the bacterial composition in the culture is initially the colonic flora isolated from the infected human host. Upon further passage, the bacterial populations may change and whether certain bacterial species predominate is not known. It has been observed for some recently axenized E. histolytica clinical isolates that bacterial flora, when provided exogenously, is beneficial to inducing encystation (Mukherjee et al., 2008). Furthermore, it has also recently been demonstrated that there is differential DNA content in parasites grown as long-term axenic cultures compared with those grown under xenic conditions (Mukherjee et al., 2008). Under axenic conditions, parasites have heterogeneous DNA content that is ~ 10-fold higher than that of xenic cultures. Re-association of an axenic isolate with bacteria reduced the DNA content to levels comparable with those of xenic cultures. Substantial changes in DNA content were also noted in E. invadens during development. The mechanisms by which bacterial association is regulating Entamoeba DNA content, and perhaps encystation potential, are not known but deserve further investigation.
2.4. Control of E. histolytica gene expression during development
Stage conversion is associated with a dramatic change in gene expression in a number of parasitic protists (Mowatt et al., 1995; Cleary et al., 2002; Bozdech et al., 2003; Ehrenkaufer et al., 2007b; Saxena et al., 2007). The changes in gene expression can be mediated by a variety of mechanisms including genetic control and transcription factor occupancy (as occurs in Giardia lamblia and a developmentally regulated Myb-domain gene) (Sun et al., 2002) or epigenetic control and histone modification (as occurs in Toxoplasma gondii in the control of tachyzoite and bradyzoite genes) (Saksouk et al., 2005).
2.4.1. Genetic control of Entamoeba development
Promoter architecture of E. histolytica genes is well described and a number of amebic transcription factors have been characterized (Singh et al., 1997; Gilchrist et al., 2001; Schaenman et al., 2001). However, this work has until recently exclusively focused on genes expressed in trophozoites. Recently, an E. histolytica myb gene that is developmentally regulated (Ehmyb-dr) and which regulates expression of a number of stage-specific genes has been identified and characterized (Ehrenkaufer et al., unpublished data). The EhMYB-dr is classified as a SHAQKY family Myb protein (Fukuzawa et al., 2006). Over-expression of the EhMYB-dr in E. histolytica trophozoites results in parasites that have a transcriptional profile, which overlaps significantly with amebic cysts. Analysis of the promoter regions of genes co-regulated by EhMYB-dr identified conserved promoter motifs. Using electrophoretic mobility shift assays, a promoter motif (CCCCCC) was identified to which protein(s) from nuclear extract from EhMYB-dr over-expressing parasites bind specifically. Further analysis using chromatin immunoprecipitation (ChIP) indicates that the EhMYB-dr binds directly to the CCCCCC promoter motif. The EhMYB-dr binding domain is reminiscent of the DNA binding domain of the Dictyostelium protein MYB-E, another member of the SHAQKY family of Myb proteins, which was implicated in the regulation of pre-stalk genes (Fukuzawa et al., 2006). We believe this work is the first identification of a developmentally regulated transcription factor in E. histolytica. A number of other potential transcription factors were identified as being developmentally regulated in the cyst microarray studies and future studies will enable further dissection of the regulatory networks that control stage conversion in E. histolytica.
2.4.2. Epigenetic control of Entamoeba development
Gene expression in E. histolytica is modulated by a number of epigenetic mechanisms including DNA methylation (Lavi et al., 2006; Ali et al., 2007) and histone modifications (Ramakrishnan et al., 2004; Isakov et al., 2008). Histone modification in E. invadens is linked to stage conversion as shown by effects of Trichostatin A (TSA) (an inhibitor of histone deacetylase) and SCFAs, both of which block encystation (Byers et al., 2005). In E. histolytica TSA has effects on histone H4 acetylation (hyperacetylation), with SCFA having the opposite effects (hypoacetylation) (Byers and Eichinger, 2008).
Recently data obtained from expression profiling linked histone acetylation to gene expression relevant to Entamoeba development. Entamoeba histolytica 200:NIH parasites were exposed to SCFA and TSA and effects on gene expression determined. Despite the changes in histone profiles and parasite growth rates induced by SCFA, there were minimal changes in amebic gene expression with ~ 0.1% of genes modulated significantly by SCFA exposure (Ehrenkaufer et al., 2007a). Thus, although E. histolytica 200:NIH trophozoites grown with SCFAs develop hypoacetylated H4 histones (Byers and Eichinger, 2008), these changes are not associated with significant alterations in the parasites’ gene expression profile. These data are in contrast to data from other eukaryotic systems in which SCFAs regulate gene expression (Fusunyan et al., 1999).
In contrast, treatment of E. histolytica trophozoites with TSA induced significant transcriptional differences (Ehrenkaufer et al., 2007a; Isakov et al., 2008). Ehrenkaufer et al. (2007) used the E. histolytica 200:NIH parasite strain and high doses of TSA (150 nM) and found decreases in parasite growth rates and transcriptional changes reminiscent of stage conversion. A total of 163 genes were regulated with 122 genes up-regulated (73 of which were also up-regulated in cysts) and 41 genes down-regulated (15 of which were also down-regulated in cysts) (Ehrenkaufer et al., 2007a). Decreased expression of a number of virulence genes was also identified. Overall, the decreased rates of growth and decreased expression of virulence genes was concordant with the expression data linking effects of TSA exposure to stage conversion. Contrasting results were obtained by Isakov et al. (2008) who identified that TSA effects on the HM-1:IMSS strain caused an increase in virulence and no induction of stage-specific genes. Whether the differences in the amebic strains or TSA doses used in each study contributed to these disparate data sets is not known and will need further investigation.
Interestingly, when the transcriptional analysis of E. histolytica cysts was performed, it was noted that there were genomic regions in which there was clustering of stage-specific genes, indicating that these genes may be co-regulated using a common mechanism (Ehrenkaufer et al., 2007b). The analysis of TSA effects on E. histolytica indicates that at least some of these genomic clusters may be controlled by histone modifications (Ehrenkaufer et al., 2007a). Whether the effect is a direct or indirect effect of histone acetylation remains to be determined.
2.4.3. Other mechanisms of gene expression regulation during Entamoeba development
Other mechanisms likely control expression of a subset of amebic stage-specific genes. One such mechanism may be via small, non-coding RNAs of the small interfering RNA (siRNA) class. There is an extensive dataset published on siRNAs, including roles of these RNA species in developmental control (Ambros et al., 2003; Rajagopalan et al., 2006). It has previously been noted that a number of genes of the RNA interference (RNAi) pathway are present in the E. histolytica genome (Ullu et al., 2004) and that siRNAs, double-stranded RNAs (dsRNAs) and short hairpin RNAs can mediate gene silencing in E. histolytica (Kaur and Lohia, 2004; Vayssie et al., 2004; Boettner et al., 2008). It has recently been identified that Entamoeba has a complex repertoire of small RNAs, with the most prevalent population of 27 nucleotides (H. Zhang, G. Ehrenkaufer, U. Singh, unpublished data). Cloning of the 27 nucleotide small RNAs identified that a substantial portion (~ 20%) of these map antisense to coding regions. Interestingly, the genes to which the antisense small RNAs map are not expressed under trophozoite conditions, the stage from which the small RNAs were cloned. However, a number of these genes are expressed in other E. histolytica strains, including strains that contain cysts. Whether the small RNAs are silencing a subset of genes relevant to amebic development is not currently known but will be an important avenue of future investigation.
2.4.5. Identification of a stress-induced gene family, Ehssp, with complex promoter regulation
A large gene family, responsive to heat and oxidative stress, was previously identified in E. histolytica (Satish et al., 2003). The cyst transcriptome indicated that a large subset of this family is also regulated in E. histolytica cysts (Ehrenkaufer et al., 2007b). In order to determine whether these co-regulated genes were coordinately controlled by promoter motifs, Hackney et al. (2007) used a Bayesian analysis algorithm. The work identified a complex transcriptional control pattern for the Ehssp gene family that was composed of three promoter motifs (GAATGATG, AACTATTTAAACATC/TC, and TGAACTTATAAACATC). Ehssp genes which had two or more of these promoter motifs in their promoters had low baseline expression and were highly up-regulated during heat shock. In contrast, genes whose promoters contained only one of the conserved promoter motifs did not respond to stress conditions. Using electrophoretic mobility shift assays (EMSA), Hackney demonstrated that amebic nuclear protein(s) bound specifically to the promoter motifs under stress conditions. This work demonstrates the ability of bioinformatic approaches to identify complex regulatory networks controlling developmentally regulated genes.
3. Future directions
Genome sequencing of E. histolytica and E. invadens has provided new information relevant to studying amebic development. The genome sequences allow a detailed comparison between the two species and have allowed whole-genome functional studies of E. histolytica; similar studies with E. invadens are underway. A high priority goal for the Entamoeba community remains the development of a high-level regulated in vitro encystation system in E. histolytica. Success in that arena, combined with genetic analysis, will allow researchers to begin to dissect the molecular networks that regulate E. histolytica development.
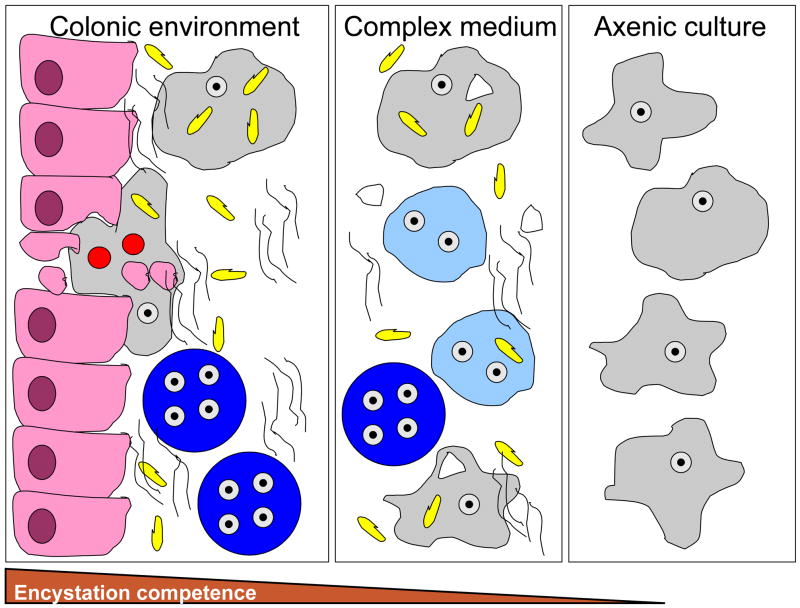
Fig. 1.
Schematic of Entamoeba histolytica in the colonic environment, in LYSGM complex xenic medium, and in axenic medium and the associated encystation ability of the parasites under each condition. Parasites in the colonic environment are exposed to colonic epithelial cells, bacteria, mucin, and red blood cells and are fully encystation competent. Parasites in complex medium are exposed to bacteria, mucin and rice starch and have some ability to encyst. Parasites in axenic culture, without any associated components as listed above, have not been shown to be encystation competent. Epithelial cell=  ; Bacteria=
; Bacteria=  ; Mucin=
; Mucin=  ; Rice starch=
; Rice starch=  ; Red blood cells=
; Red blood cells=  ; E. histolytica trophozoites=
; E. histolytica trophozoites=  ; Immature E. histolytica cysts=
; Immature E. histolytica cysts=  ; Mature E. histolytica cysts=
; Mature E. histolytica cysts=  .
.
Fig. 2.
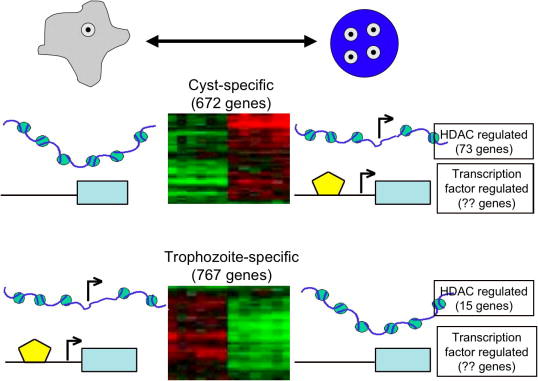
Two mechanisms by which genetic control of development is regulated. Cyst and trophozoite-specific genes in Entamoeba histolytica can be regulated by chromatin structure or by promoter factor occupancy. Of the developmentally regulated genes, 73 cyst-specific genes are regulated by histone deacetylase inhibitors (HDAC) and 15 trophozoite-specific genes are regulated by HDAC. The number of genes in each category that are regulated by promoter occupancy is unknown. Entamoeba histolytica trophozoite=  ; E. histolytica cyst=
; E. histolytica cyst=  ; Microarray expression data--green (no expression) and red (expressed); DNA wrapped around a histone core =
; Microarray expression data--green (no expression) and red (expressed); DNA wrapped around a histone core =  ; transcription factor=
; transcription factor=  ; Expressed gene: bent arrow.
; Expressed gene: bent arrow.
Acknowledgments
We gratefully acknowledge all members of the Singh laboratory for helpful comments and suggestions. We thank C. Graham Clark for sharing unpublished data and for providing valuable insights. US and GME were supported in part from NIH grants AI-069382 and AI-068899.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali IK, Ehrenkaufer GM, Hackney JA, Singh U. Growth of the protozoan parasite Entamoeba histolytica in 5-azacytidine has limited effects on parasite gene expression. BMC genomics. 2007;8:7. doi: 10.1186/1471-2164-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Bailey GB, Rengypian S. Osmotic stress as a factor controlling encystation of Entamoeba invadens. Arch Invest Med (Mex) 1980;11:11–16. [PubMed] [Google Scholar]
- Balamuth W. Biological studies on Entamoeba histolytica. III. Induced encystation in several mediums, including an account of a new procedure. J Inf Dis. 1951;88:230–236. doi: 10.1093/infdis/88.3.230. [DOI] [PubMed] [Google Scholar]
- Balamuth W, Wieboldt ML. Comparative growth cycles of Entamoeba histolytica with different combinations of bacteria. Am J Trop Med Hyg. 1951;31:192–205. doi: 10.4269/ajtmh.1951.s1-31.192. [DOI] [PubMed] [Google Scholar]
- Barron-Gonzalez MP, Villarreal-Trevino L, Resendez-Perez D, Mata-Cardenas BD, Morales-Vallarta MR. Entamoeba histolytica: cyst-like structures in vitro induction. Exp Parasitol. 2008;118:600–603. doi: 10.1016/j.exppara.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Boettner DR, Huston CD, Linford AS, Buss SN, Houpt E, Sherman NE, Petri WA., Jr Entamoeba histolytica phagocytosis of human erythrocytes involves PATMK, a member of the transmembrane kinase family. PLoS pathogens. 2008;4:e8. doi: 10.1371/journal.ppat.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z, Zhu J, Joachimiak MP, Cohen FE, Pulliam B, DeRisi JL. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome biology. 2003;4:R9. doi: 10.1186/gb-2003-4-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers J, Faigle W, Eichinger D. Colonic short-chain fatty acids inhibit encystation of Entamoeba invadens. Cell Microbiol. 2005;7:269–279. doi: 10.1111/j.1462-5822.2004.00457.x. [DOI] [PubMed] [Google Scholar]
- Byers J, Eichinger D. Acetylation of the Entamoeba histone H4 N-terminal domain is influenced by short-chain fatty acids that enter trophozoites in a pH-dependent manner. Int J Parasitol. 2008;38:57–64. doi: 10.1016/j.ijpara.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chayen A, Avron B, Nuchamowitz Y, Mirelman D. Appearance of sialoglycoproteins in encysting cells of Entamoeba histolytica. Infect Immun. 1988;56:673–681. doi: 10.1128/iai.56.3.673-681.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Cleary MD, Singh U, Blader IJ, Brewer JL, Boothroyd JC. Toxoplasma gondii asexual development: identification of developmentally regulated genes and distinct patterns of gene expression. Eukaryot Cell. 2002;1:329–340. doi: 10.1128/EC.1.3.329-340.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppi A, Merali S, Eichinger D. The enteric parasite Entamoeba uses an autocrine catecholamine system during differentiation into the infectious cyst stage. J Biol Chem. 2002;277:8083–8090. doi: 10.1074/jbc.M111895200. [DOI] [PubMed] [Google Scholar]
- Das S, Gillin FD. Chitin synthase in encysting Entamoeba invadens. Biochem J. 1991;280 (Pt 3):641–647. doi: 10.1042/bj2800641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Van Dellen K, Bulik D, Magnelli P, Cui J, Head J, Robbins PW, Samuelson J. The cyst wall of Entamoeba invadens contains chitosan (deacetylated chitin) Mol Biochem Parasitol. 2006;148:86–92. doi: 10.1016/j.molbiopara.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Davis PH, Schulze J, Stanley SL., Jr Transcriptomic comparison of two Entamoeba histolytica strains with defined virulence phenotypes identifies new virulence factor candidates and key differences in the expression patterns of cysteine proteases, lectin light chains, and calmodulin. Mol Biochem Parasitol. 2007;151:118–128. doi: 10.1016/j.molbiopara.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Diamond LS. Axenic cultivation of Entamoeba histolytica. Science. 1961;134:336–337. doi: 10.1126/science.134.3475.336. [DOI] [PubMed] [Google Scholar]
- Dobell C. Researches on the intestinal protozoa of monkeys and man. Parasitology. 1928;20:357–412. doi: 10.1017/s0031182000084225. [DOI] [PubMed] [Google Scholar]
- Dumur CI, Garrett CT, Archer KJ, Nasim S, Wilkinson DS, Ferreira-Gonzalez A. Evaluation of a linear amplification method for small samples used on high-density oligonucleotide microarray analysis. Analytical biochemistry. 2004;331:314–321. doi: 10.1016/j.ab.2004.03.040. [DOI] [PubMed] [Google Scholar]
- Ehrenkaufer GM, Eichinger DJ, Singh U. Trichostatin A effects on gene expression in the protozoan parasite Entamoeba histolytica. BMC genomics. 2007a;8:216. doi: 10.1186/1471-2164-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenkaufer GM, Haque R, Hackney JA, Eichinger DJ, Singh U. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechanisms of stage conversion in a protozoan parasite. Cell Microbiol. 2007b;9:1426–1444. doi: 10.1111/j.1462-5822.2006.00882.x. [DOI] [PubMed] [Google Scholar]
- Eichinger D. Encystation of Entamoeba parasites. Bioessays. 1997;19:633–639. doi: 10.1002/bies.950190714. [DOI] [PubMed] [Google Scholar]
- Eichinger D. A role for a galactose lectin and its ligands during encystment of Entamoeba. J Eukaryot Microbiol. 2001;48:17–21. doi: 10.1111/j.1550-7408.2001.tb00411.x. [DOI] [PubMed] [Google Scholar]
- Frisardi M, Ghosh SK, Field J, Van Dellen K, Rogers R, Robbins P, Samuelson J. The most abundant glycoprotein of amebic cyst walls (Jacob) is a lectin with five Cys-rich, chitin-binding domains. Infect Immun. 2000;68:4217–4224. doi: 10.1128/iai.68.7.4217-4224.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa M, Zhukovskaya NV, Yamada Y, Araki T, Williams JG. Regulation of Dictyostelium prestalk-specific gene expression by a SHAQKY family MYB transcription factor. Development. 2006;133:1715–1724. doi: 10.1242/dev.02327. [DOI] [PubMed] [Google Scholar]
- Fusunyan RD, Quinn JJ, Fujimoto M, MacDermott RP, Sanderson IR. Butyrate switches the pattern of chemokine secretion by intestinal epithelial cells through histone acetylation. Molecular medicine. 1999;5:631–640. [PMC free article] [PubMed] [Google Scholar]
- Gangopadhyay SS, Ray SS, Kennady K, Pande G, Lohia A. Heterogeneity of DNA content and expression of cell cycle genes in axenically growing Entamoeba histolytica HM1:IMSS clone A. Mol Biochem Parasitol. 1997;90:9–20. doi: 10.1016/s0166-6851(97)00156-4. [DOI] [PubMed] [Google Scholar]
- Gilchrist CA, Holm CF, Hughes MA, Schaenman JM, Mann BJ, Petri WA., Jr Identification and characterization of an Entamoeba histolytica upstream regulatory element 3 sequence-specific DNA-binding protein containing EF-hand motifs. J Biol Chem. 2001;276:11838–11843. doi: 10.1074/jbc.M007375200. [DOI] [PubMed] [Google Scholar]
- Gilchrist CA, Houpt E, Trapaidze N, Fei Z, Crasta O, Asgharpour A, Evans C, Martino-Catt S, Baba DJ, Stroup S, Hamano S, Ehrenkaufer G, Okada M, Singh U, Nozaki T, Mann BJ, Petri WA., Jr Impact of intestinal colonization and invasion on the Entamoeba histolytica transcriptome. Mol Biochem Parasitol. 2006;147:163–176. doi: 10.1016/j.molbiopara.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Gonzalez J, Bai G, Frevert U, Corey EJ, Eichinger D. Proteasome-dependent cyst formation and stage-specific ubiquitin mRNA accumulation in Entamoeba invadens. Eur J Biochem. 1999;264:897–904. doi: 10.1046/j.1432-1327.1999.00682.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Salazar F, Viader-Salvado JM, Martinez-Rodriguez HG, Campos-Gongora E, Mata-Cardenas BD, Said-Fernandez S. Identification of seven chemical factors that favor high-quality Entamoeba histolytica cyst-like structure formation under axenic conditions. Arch Med Res. 2000;31:S192–193. doi: 10.1016/s0188-4409(00)00193-4. [DOI] [PubMed] [Google Scholar]
- Hackney JA, Ehrenkaufer GM, Singh U. Identification of putative transcriptional regulatory networks in Entamoeba histolytica using Bayesian inference. Nucleic acids research. 2007;35:2141–2152. doi: 10.1093/nar/gkm028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque R, Ali IM, Petri WA., Jr Prevalence and immune response to Entamoeba histolytica infection in preschool children in Bangladesh. Am J Trop Med Hyg. 1999;60:1031–1034. doi: 10.4269/ajtmh.1999.60.1031. [DOI] [PubMed] [Google Scholar]
- Haque R, Huston CD, Hughes M, Houpt E, Petri WA., Jr Amebiasis. N Engl J Med. 2003;348:1565–1573. doi: 10.1056/NEJMra022710. [DOI] [PubMed] [Google Scholar]
- Isakov E, Siman-Tov R, Weber C, Guillen N, Ankri S. Trichostatin A regulates peroxiredoxin expression and virulence of the parasite Entamoeba histolytica. Mol Biochem Parasitol. 2008;158:82–94. doi: 10.1016/j.molbiopara.2007.11.014. [DOI] [PubMed] [Google Scholar]
- Jensen JB. In vitro cultivation of protozoan parasites. CRC Press; Boca Raton, Fla: 1983. [Google Scholar]
- Kaur G, Lohia A. Inhibition of gene expression with double strand RNA interference in Entamoeba histolytica. Biochem Biophys Res Commun. 2004;320:1118–1122. doi: 10.1016/j.bbrc.2004.06.064. [DOI] [PubMed] [Google Scholar]
- Lavi T, Isakov E, Harony H, Fisher O, Siman-Tov R, Ankri S. Sensing DNA methylation in the protozoan parasite Entamoeba histolytica. Mol Microbiol. 2006;62:1373–1386. doi: 10.1111/j.1365-2958.2006.05464.x. [DOI] [PubMed] [Google Scholar]
- Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, Nozaki T, Suh B, Pop M, Duchene M, Ackers J, Tannich E, Leippe M, Hofer M, Bruchhaus I, Willhoeft U, Bhattacharya A, Chillingworth T, Churcher C, Hance Z, Harris B, Harris D, Jagels K, Moule S, Mungall K, Ormond D, Squares R, Whitehead S, Quail MA, Rabbinowitsch E, Norbertczak H, Price C, Wang Z, Guillen N, Gilchrist C, Stroup SE, Bhattacharya S, Lohia A, Foster PG, Sicheritz-Ponten T, Weber C, Singh U, Mukherjee C, El-Sayed NM, Petri WA, Jr, Clark CG, Embley TM, Barrell B, Fraser CM, Hall N. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- MacFarlane RC, Singh U. Identification of differentially expressed genes in virulent and nonvirulent Entamoeba species: potential implications for amebic pathogenesis. Infect Immun. 2006;74:340–351. doi: 10.1128/IAI.74.1.340-351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makioka A, Kumagai M, Kobayashi S, Takeuchi T. Possible role of calcium ions, calcium channels and calmodulin in excystation and metacystic development of Entamoeba invadens. Parasitol Res. 2002a;88:837–843. doi: 10.1007/s00436-002-0676-6. [DOI] [PubMed] [Google Scholar]
- Makioka A, Kumagai M, Kobayashi S, Takeuchi T. Inhibition of excystation and metacystic development of Entamoeba invadens by the dinitroaniline herbicide oryzalin. J Parasitol. 2002b;88:994–999. doi: 10.1645/0022-3395(2002)088[0994:IOEAMD]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Makioka A, Kumagai M, Kobayashi S, Takeuchi T. Involvement of signaling through protein kinase C and phosphatidylinositol 3-kinase in the excystation and metacystic development of Entamoeba invadens. Parasitol Res. 2003;91:204–208. doi: 10.1007/s00436-003-0955-x. [DOI] [PubMed] [Google Scholar]
- Makioka A, Kumagai M, Kobayashi S, Takeuchi T. Entamoeba invadens: cysteine protease inhibitors block excystation and metacystic development. Exp Parasitol. 2005;109:27–32. doi: 10.1016/j.exppara.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Makioka A, Kumagai M, Kobayashi S, Takeuchi T. Effect of artificial gastrointestinal fluids on the excystation and metacystic development of Entamoeba invadens. Parasitol Res. 2006;98:443–446. doi: 10.1007/s00436-005-0025-7. [DOI] [PubMed] [Google Scholar]
- Mowatt MR, Lujan HD, Cotten DB, Bowers B, Yee J, Nash TE, Stibbs HH. Developmentally regulated expression of a Giardia lamblia cyst wall protein gene. Mol Microbiol. 1995;15:955–963. doi: 10.1111/j.1365-2958.1995.tb02364.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee C, Clark CG, Lohia A. Entamoeba shows reversible variation in ploidy and under different growth conditions and between life cycle phases. PLoS Negl Trop Dis. 2008;2(8):e281. doi: 10.1371/journal.pntd.0000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picazarri K, Nakada-Tsukui K, Nozaki T. Autophagy during proliferation and encystation in the protozoan parasite Entamoeba invadens. Infect Immun. 2008;76:278–288. doi: 10.1128/IAI.00636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & development. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan G, Gilchrist CA, Musa H, Torok MS, Grant PA, Mann BJ, Petri WA., Jr Histone acetyltransferases and deacetylase in Entamoeba histolytica. Mol Biochem Parasitol. 2004;138:205–216. doi: 10.1016/j.molbiopara.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Robinson GL. The laboratory diagnosis of human parasitic amoebae. Trans R Soc Trop Med Hyg. 1968;62:285–294. doi: 10.1016/0035-9203(68)90170-3. [DOI] [PubMed] [Google Scholar]
- Saksouk N, Bhatti MM, Kieffer S, Smith AT, Musset K, Garin J, Sullivan WJ, Jr, Cesbron-Delauw MF, Hakimi MA. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Molecular and cellular biology. 2005;25:10301–10314. doi: 10.1128/MCB.25.23.10301-10314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez L, Enea V, Eichinger D. Identification of a developmentally regulated transcript expressed during encystation of Entamoeba invadens. Mol Biochem Parasitol. 1994;67:125–135. doi: 10.1016/0166-6851(94)90102-3. [DOI] [PubMed] [Google Scholar]
- Santi-Rocca J, Weber C, Guigon G, Sismeiro O, Coppee JY, Guillen N. The lysine-and glutamic acid-rich protein KERP1 plays a role in Entamoeba histolytica liver abscess pathogenesis. Cell Microbiol. 2008;10:202–217. doi: 10.1111/j.1462-5822.2007.01030.x. [DOI] [PubMed] [Google Scholar]
- Satish S, Bakre AA, Bhattacharya S, Bhattacharya A. Stress-dependent expression of a polymorphic, charged antigen in the protozoan parasite Entamoeba histolytica. Infect Immun. 2003;71:4472–4486. doi: 10.1128/IAI.71.8.4472-4486.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena A, Lahav T, Holland N, Aggarwal G, Anupama A, Huang Y, Volpin H, Myler PJ, Zilberstein D. Analysis of the Leishmania donovani transcriptome reveals an ordered progression of transient and permanent changes in gene expression during differentiation. Mol Biochem Parasitol. 2007;152:53–65. doi: 10.1016/j.molbiopara.2006.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaenman JM, Gilchrist CA, Mann BJ, Petri WA., Jr Identification of two Entamoeba histolytica sequence-specific URE4 enhancer-binding proteins with homology to the RNA-binding motif RRM. J Biol Chem. 2001;276:1602–1609. doi: 10.1074/jbc.M006866200. [DOI] [PubMed] [Google Scholar]
- Singh U, Rogers JB, Mann BJ, Petri WA., Jr Transcription initiation is controlled by three core promoter elements in the hgl5 gene of the protozoan parasite Entamoeba histolytica. Proc Natl Acad Sci U S A. 1997;94:8812–8817. doi: 10.1073/pnas.94.16.8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SL., Jr Amoebiasis. Lancet. 2003;361:1025–1034. doi: 10.1016/S0140-6736(03)12830-9. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Hamblin K, Perez-Brocal V, Gaston D, Richmond GS, van der Giezen M, Clark CG, Roger AJ. Organelles in Blastocystis that blur the distinction between mitochondria and hydrogenosomes. Curr Biol. 2008;18:580–585. doi: 10.1016/j.cub.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CH, Palm D, McArthur AG, Svard SG, Gillin FD. A novel Myb-related protein involved in transcriptional activation of encystation genes in Giardia lamblia. Mol Microbiol. 2002;46:971–984. doi: 10.1046/j.1365-2958.2002.03233.x. [DOI] [PubMed] [Google Scholar]
- Turner NA, Eichinger D. Entamoeba invadens: the requirement for galactose ligands during encystment. Exp Parasitol. 2007;116:467–474. doi: 10.1016/j.exppara.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E, Tschudi C, Chakraborty T. RNA interference in protozoan parasites. Cell Microbiol. 2004;6:509–519. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- Van Dellen KL, Bulik DA, Specht CA, Robbins PW, Samuelson JC. Heterologous expression of an Entamoeba histolytica chitin synthase in Saccharomyces cerevisiae. Eukaryot Cell. 2006a;5:203–206. doi: 10.1128/EC.5.1.203-206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dellen KL, Chatterjee A, Ratner DM, Magnelli PE, Cipollo JF, Steffen M, Robbins PW, Samuelson J. Unique posttranslational modifications of chitin-binding lectins of Entamoeba invadens cyst walls. Eukaryot Cell. 2006b;5:836–848. doi: 10.1128/EC.5.5.836-848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vayssie L, Vargas M, Weber C, Guillen N. Double-stranded RNA mediates homology-dependent gene silencing of gamma-tubulin in the human parasite Entamoeba histolytica. Mol Biochem Parasitol. 2004;138:21–28. doi: 10.1016/j.molbiopara.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Vazquezdelara-Cisneros LG, Arroyo-Begovich A. Induction of encystation of Entamoeba invadens by removal of glucose from the culture medium. J Parasitol. 1984;70:629–633. [PubMed] [Google Scholar]
- Villagomez-Castro JC, Calvo-Mendez C, Lopez-Romero E. Chitinase activity in encysting Entamoeba invadens and its inhibition by allosamidin. Mol Biochem Parasitol. 1992;52:53–62. doi: 10.1016/0166-6851(92)90035-i. [DOI] [PubMed] [Google Scholar]
- Wang Z, Samuelson J, Clark CG, Eichinger D, Paul J, Van Dellen K, Hall N, Anderson I, Loftus B. Gene discovery in the Entamoeba invadens genome. Mol Biochem Parasitol. 2003;129:23–31. doi: 10.1016/s0166-6851(03)00073-2. [DOI] [PubMed] [Google Scholar]
- Weber C, Guigon G, Bouchier C, Frangeul L, Moreira S, Sismeiro O, Gouyette C, Mirelman D, Coppee JY, Guillen N. Stress by heat shock induces massive down regulation of genes and allows differential allelic expression of the Gal/GalNAc lectin in Entamoeba histolytica. Eukaryot Cell. 2006;5:871–875. doi: 10.1128/EC.5.5.871-875.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Amoebiasis. Wkly Epidemiol Rec. 1997;72:97–99. [PubMed] [Google Scholar]