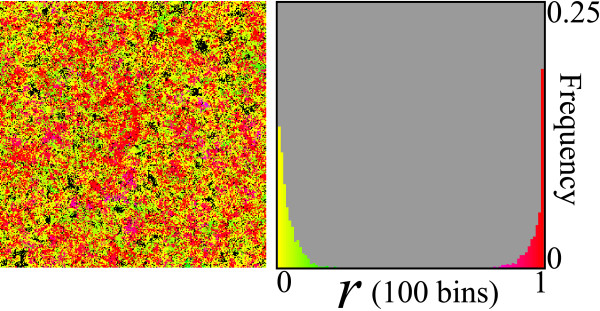
Figure 9.
Speciation: emergence of replicase transcribers (r ≈ 1) and parasitic genome copiers (r ≈ 0). This figure shows the results of a simulation where kSM and kSP can also evolve the correlation with kDM and kDP is not presumed, but it may evolve. The right panel shows a population distribution of r. The abscissa is r in the range of [0,1] with 100 bins. The coordinate is the frequency of individuals in the range of [0,0.25]. A bimodal distribution indicates a speciation-like phenomenon [the population distribution of kSP/(kSP + kSM) also shows a similar bimodal distribution; data not shown]. The left panel shows the spatial distribution of individuals colored by value of r. The color coding is the same as in the right panel (note that the color coding is different from that of Fig. 2). For the sake of comparison, the size of the grid is set to 300 × 300 as in Fig. 2. The parameters are as follows: 0.5(kSM + kSP) = (kDM + kDP) = 1; Δ = 0.01; d = 0.01; μ = 0.01; δr = 0.1. The mutation of kSM and kSP is implemented in the same way as that of kDM and kDP. [Additionaly, we observed the speciation-like phenomenon also in the following parameter conditions: d = 0.001 and 0.1 ≤ Δ ≤ 0.32; d = 0.01 and 0.001 ≤ Δ ≤ 0.1; d = 0.02 and Δ = 0.1.].