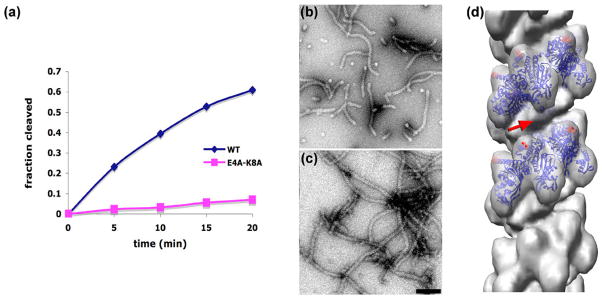
Figure 4.
RecA residues E4 and K8 are directly involved in the coordination and cleavage of cI. The rate of cI93–236 cleavage is greatly reduced when the RecA-E4A-K8A mutant is used in comparison to wt RecA (a). The repeating GTG 48-mer has been used along with ATP-γ-S, as in Fig. 1. An electron micrograph of RecA-E4A-K8A polymerized on the repeating GTG 48-mer (b) shows a large degree of end-to-end stacking of these 48-mers, as the filaments are considerably longer than the ~ 2.5 helical turns that would be expected in the absence of stacking. When the non-cleavable cI101–229TM fragment is added to these RecA-E4A-K8A filaments (c) there are no dramatic differences. A three-dimensional reconstruction (d), generated from 3,980 segments of filaments as shown in (c), lacks the bridge of density that is seen with the wt RecA protein under the same conditions when the cI101–229TM fragment is bound (red arrow, Fig. 3f). Residues E4 and K8 are marked as red spheres in (d). The scale bar (c) is 1,000 Å.