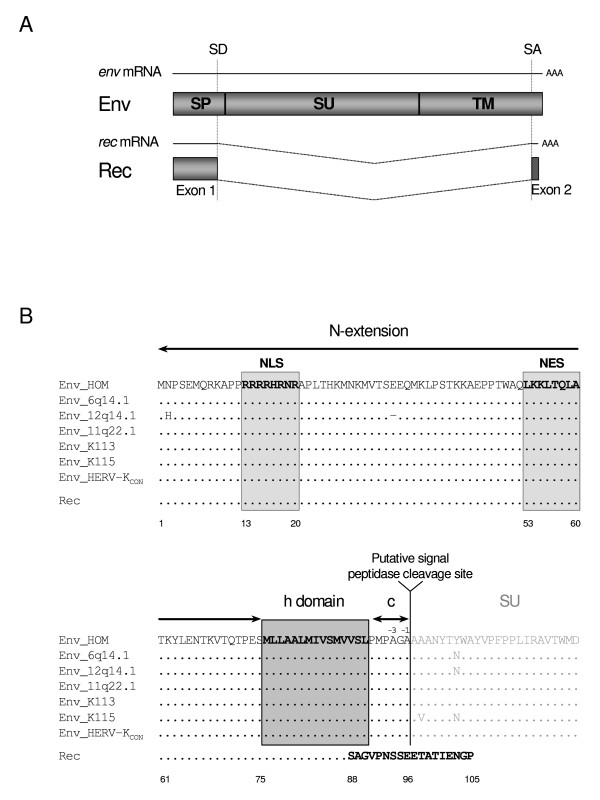
Figure 2.
Comparison of HERV-K(HML-2) SP and Rec sequences. (A) env mRNA encodes an Env precursor protein that is cleaved in the ER by signal peptidase releasing SP. In the Golgi, the Env precursor is further processed and cleaved by a furin-like endoprotease to give rise to surface (SU) and transmembrane (TM) subunits. rec mRNA is a splice product of env mRNA and encodes Rec. The first exon of Rec overlaps with SP while the second exon is translated from a different reading frame. SD/SA: rec splice donor and acceptor sites. (B) SP and Rec amino acid sequence alignment. The human genome contains six proviruses with complete Env ORFs [13]. HERV-K(HML-2.HOM) is an almost intact provirus located on chromosome 7p22.1 (Env_HOM) [60]. Chromosomal localizations of other Env encoding loci are indicated. HERV-K113 (Env_K113) and HERV-K115 (Env_K115) are two polymorphic proviruses located on chromosomes 19p12 and 8p23.1, respectively [4]. The alignment also includes HERV-KCON, a recently engineered "infectious" provirus [12], and the Rec sequence [27]. Rec exon 1 (aa 1–87) is also found in SP while the second exon of Rec (aa 88–105) is translated from a different reading frame. N: N-extension (aa 1–75); h: hydrophobic h domain (aa 76–90); C: polar domain (aa 91–96); -3,-1:position of small uncharged residues. By analogy with motifs previously characterized in Rec, a putative arginine-rich nuclear localization signal (NLS; aa 13–20) and a leucine-rich nuclear export signal (NES; aa 54–60) are present in HML-2 SP.