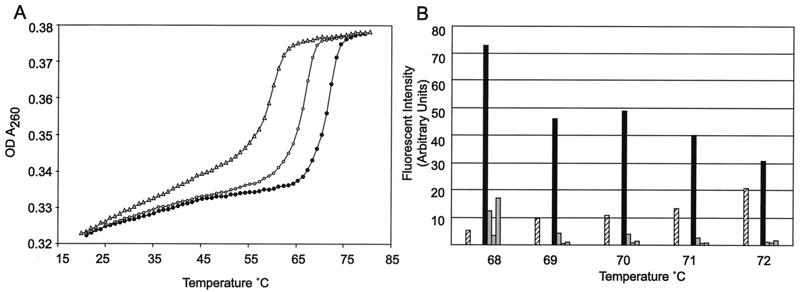
Figure 3.
Melting curve of tags with complements. (A) Approximately 0.35 OD units of each double-stranded oligonucleotide was resuspended in 10 mM sodium phosphate, pH 7.6/50 mM NaCl/3 mM MgCl2. The samples were heated at 1°C per min, and the OD at 260 nm was measured and recorded. Reactions consisted of one common oligonucleotide, cccatcactttatcaatcaacatatcacaaaaatctcc, and a second oligonucleotide as follows: perfect match, (●) ggagatttttgtgatatgttgattgataaagtgatggg; one-word mismatch, (○) ggatgattttgtgatatgttgattgataaagtgatggg; two-word mismatch, (▵) ggagatttttgtttgatgttttgtgataaagtgatggg. (B) Microbeads bearing the oligonucleotide ggagatttttgtgatatgttgattgataaagtgatggg were loaded with FAM-labeled cDNA tagged with this sequence. In separate tubes, three cDNAs whose tags differed at the first, third, or fifth positions also were loaded. The samples then were washed in 50 mM Tris/50 mM NaCl/3 mM Mg2Cl at increasing temperatures as indicated. Microbeads then were analyzed by FACS, and the means of fluorescence intensities were plotted for the perfect match of the tag (filled bar), the mismatches (gray bars), and the ratios (cross-hatched bar) of the perfect signal to the average noise of the three mismatches.