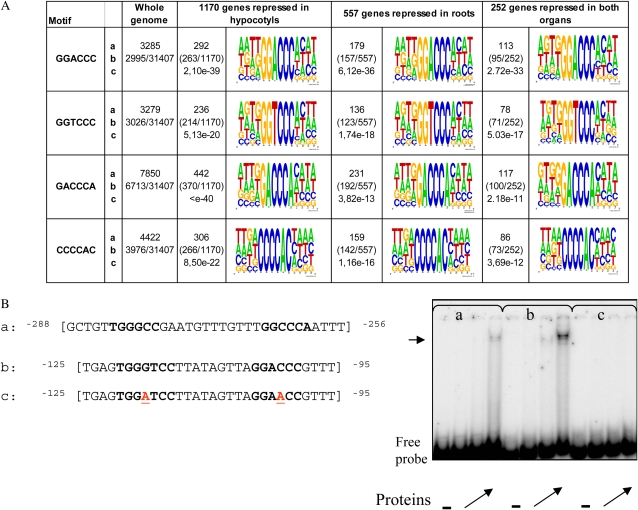
Figure 7.
Definition of putative motifs in the query set of promoter sequences of genes repressed in hypocotyls, in roots, or in both organs according to the Motif Analysis program at TAIR and test of binding activity to MBP-TCP20. A, Sequence logos of the overrepresented motifs found in the AtTCP20∷EAR-repressed genes: a, absolute number of motifs in the query set; b, number of promoter sequences in the query set containing a motif; c, P value for binomial distribution. The logo was created based on motif instances in the query set and surrounding bases using WebLogo (Crooks et al., 2004). The height of the symbols within the stack reflects the relative frequency of the corresponding nucleic acid at that position. B, EMSA for binding of MBP-TCP20 to class I motifs: a, sequence of the wild-type-CC probe from the AtPCNA-2 promoter used as a control; b and c, sequences of wild-type and mutated probes from the At2g36870 promoter used for EMSA as indicated. Class II and I motifs are shown in boldface, and the mutated nucleotides are underlined (in red) in the mutant probe. The − lanes at right correspond to migration of the probe without protein. Other lanes correspond to migration obtained with increasing quantities of the recombinant protein (angled arrows). The horizontal arrow indicates the complex obtained with the probe corresponding to oligonucleotides a and b. [See online article for color version of this figure.]