Abstract
During arbuscular mycorrhizal (AM) colonization, a focal accumulation of organelles occurs in root epidermal cells, prior to fungal penetration, beneath adhering hyphopodia. This is followed by the appearance of the prepenetration apparatus (PPA), a transcellular column of cytoplasm connected to the nucleus and rich in cytoskeleton and secretory endomembranes. This apparatus appears to be responsible for the construction of an apoplastic compartment that confines the fungus within the cell lumen. To identify AM-specific elements within the PPA response, we challenged root cultures of Medicago truncatula, expressing a green fluorescent protein tag for the endoplasmic reticulum, with an AM symbiont, a necrotrophic pathogen, a hemibiotrophic pathogen, a noncompatible endomycorrhizal fungus, or abiotic physical stimuli. Parallel experiments were made on a M. truncatula nonsymbiotic mutant (doesn't make infections, dmi3-1). The results have highlighted a correlation between physical stimulation of the cell surface and nuclear repositioning. Cytoplasmic aggregation was only induced by contact with compatible fungi, whereas PPA appearance was specifically triggered by the AM fungus. The dmi3-1 mutant did not develop cytoplasmic aggregation or PPA and underwent cell death upon physical stimulation. The up-regulation of an expansin-like gene, already identified as an early marker of AM fungal contact, was triggered in wild-type roots by all the fungi tested. Such observations identify responses that are specific to mycorrhizal interactions and extend the role of the DMI3 protein, a calcium/calmodulin-dependent kinase, from symbiotic to pathogenic interactions.
Cytoplasm reorganization in epidermal cells, including nuclear repositioning, is a well-documented response in several plant-microbe interactions (O'Connell and Panstruga, 2006; Genre and Bonfante, 2007; Hardham et al., 2007). Tissue wounding (Heath et al., 1997) and physical stimulation of cells (Gus-Mayer et al., 1998; Jaffe et al., 2002) also cause nuclear repositioning. Pathogenic fungi are known to trigger the so-called cytoplasmic aggregation (CA), which is a cytoskeleton-driven accumulation of organelles, including the plant nucleus, at the penetration site (Hardham et al., 2007). CA is believed to form part of the plant defense response that leads to the formation of cell wall appositions (papillae) and the localized release of defense-related compounds (Hardham et al., 2008). The plant endoplasmic reticulum (ER), Golgi bodies, and peroxisomes in fact accumulate in large quantities in advance of fungal penetration (Takemoto et al., 2003; Koh et al., 2005), suggesting the initiation of localized secretory activity. Such reactions have been described in both susceptible and resistant interactions (Heath et al., 1997), supporting the hypothesis that they may represent the cell's first defense strategy, and anticipate hypersensitive and systemic responses (Dangl and Jones, 2001; Takemoto and Hardham, 2004; Kwon et al., 2008).
Root epidermal cell response to arbuscular mycorrhizal (AM) fungi (Genre et al., 2005) may show analogies with CA-related cell reorganization. Focal accumulation of ER and nuclear repositioning take place in epidermal cells at the sites where AM fungi form hyphopodia (Genre and Bonfante, 2007). Subsequently, but prior to fungal ingress, the host cell develops a cytoplasmic column that traverses the cell in a root centripetal direction. This unique structure, the prepenetration apparatus (PPA), is apparently related to the construction of the trans-cellular apoplastic compartment where the fungus is hosted. The PPA is particularly rich in secretory membranes, ER, and cytoskeletal elements (Genre et al., 2005, 2008), and its assembly is closely associated with a second migration of the plant nucleus, as it moves toward the inner cell wall facing the root cortex. Only after the trans-cellular cytoplasmic bridge has been built across the cell does the AM fungus enter into the epidermis as a first step toward inner tissue colonization. Once the AM fungus is in the root cortex the PPA mechanism is replicated and modulated to host the arbuscules (Genre et al., 2008), the highly branched structures that represent the main site of nutrient exchanges in a fully functional symbiosis (Harrison, 2005; Paszkowski, 2006).
Taken together, these observations suggest the existence of at least partly overlapping mechanisms in the prepenetration responses to AM and pathogenic fungi, where the local accumulation of organelles and nuclear repositioning appear to be common features (O'Connell and Panstruga, 2006).
In this study, we have attempted to directly compare epidermal cell responses that anticipate AM fungal entry with those triggered by contact with other pathogenic and nonpathogenic fungi, as well as abiotic stimuli. Medicago truncatula roots were challenged with Gigaspora margarita (an AM fungus), Colletotrichum trifolii (a hemibiotrophic pathogen), Phoma medicaginis (a necrotrophic pathogen), Oidiodendron maius (an ericoid endomycorrhizal fungus), and thigmo stimulation (physical contact with a micromanupulator-controlled microneedle). While M. truncatula establishes a compatible interaction with the first three fungi—i.e. there is a successful colonization of the root, irrespectively of its beneficial or pathological outcome—this plant is a nonhost for the ericoid fungus O. maius, which in fact does not colonize the roots.
Cell responses were visualized by constitutively expressing GFP-HDEL, a GFP tag that labels the ER, in Agrobacterium rhizogenes-transformed roots of M. truncatula. The ER labeling obtained with this construct also outlines the nuclear envelope, thus providing useful information on the nucleus position. Parallel experiments were conducted with plants carrying a mutation in the DMI3 (for DOESN'T MAKE INFECTIONS) gene, a calcium and calmodulin-dependent kinase that is central in the signaling pathways that mediate AM and nodulation (Lévy et al., 2004). We have also monitored the expression levels of two M. truncatula genes showing homology with an Expansin-like B gene (Exp-like) and an Avr9-Cf9 rapidly expressed protein (ACRE264). Since these sequences were shown to be differentially regulated in a DMI3-dependent manner during PPA development (Siciliano et al., 2007), we monitored their expression level by real-time reverse transcription (RT)-PCR, to compare their potential involvement in the various the plant-fungus interactions that were examined.
The results presented here highlight correlations between: (1) physical stimulation of the cell surface and nuclear repositioning; (2) contact with compatible fungi and CA, in addition to nuclear movement; and (3) AM fungal adhesion and PPA appearance. In addition, our experiments suggest an unexpected and more general involvement of DMI3 in pathogenic interactions and thigmotropic responses, since dmi3-1 mutants did not develop CA or PPA under any of the conditions tested and underwent cell death upon physical stimulation. In contrast to ACRE264, the analysis of Exp-like expression suggests its involvement in all the root-fungus interactions examined. Lastly, the results on the M. truncatula/O. maius combination provide a first view of the cellular and molecular interactions between mycorrhizal fungi and nonhost plants.
RESULTS
G. margarita Elicits Nuclear Repositioning, CA, and PPA Formation
Root epidermal cells of wild-type cultured roots responded to contact with G. margarita with an initial nuclear repositioning under the hyphopodium (the branched and swollen hyphae that adhere to the root surface), associated with local CA, as revealed by ER fluorescence imaging (Fig. 1A, left). This was estimated to take place within about 2 h and was followed by a second nuclear migration across the cell lumen. This was associated with the assembly of the PPA, as shown in the rightmost cell of Figure 1A. Actual cell penetration by the infection hypha occurred 6 to 8 h after hyphopodium development (Fig. 2A). Such events, described in detail by Siciliano et al. (2007), are consistent with the first description of PPA elicitation by another AM symbiont, Gigaspora gigantea (Genre et al., 2005).
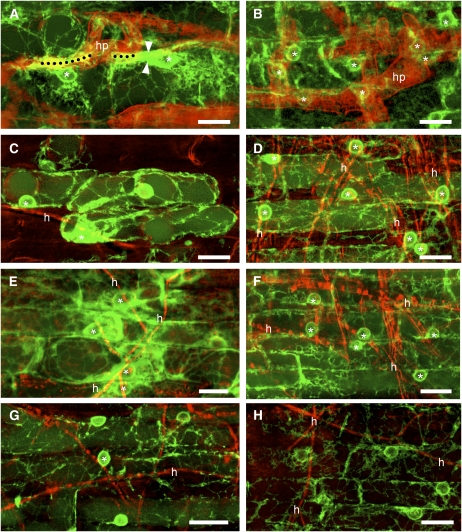
Figure 1.
Epidermal cell responses in wild-type (A, C, E, and G) and dmi3-1 (B, D, F, and H) root organ cultures of M. truncatula expressing GFP-HDEL contacted by G. margarita (A and B), C. trifolii (C and D), P. medicaginis (E and F), and O. maius (G and H). All the pictures are projections of optical sections showing GFP fluorescence (green) overlaid with inverted bright-field images false colored in red to improve the visibility of the hyphal tracks. A, Left side: Accumulation of ER and nucleus (*) repositioning beneath the contact area (dotted line) with a G. margarita hyphopodium branch. Right side: A complete PPA (arrowheads) connects the contact site (dotted line) with the nucleus (*) at the end of its transcellular migration. The apparent PPA orientation from left to right is due to the top-view projection of the confocal picture, which flattens the image depth. The PPA is actually obliquely oriented from the cell outer surface to its inner side, where the nucleus is positioned. B, G. margarita hyphopodium contact triggers nuclear repositioning, but no accumulation of ER in the dmi3-1 mutant. C, C. trifolii induces a large accumulation of ER under a hypha adhering to the epidermal cell surface. The nucleus (*) is also repositioned underneath the contact site, although it appears hidden by the overlapped ER cisternae in this projected image. D, The adhesion of C. trifolii to the surface of the dmi3-1 root instead only triggers nuclear repositioning. E, Wild-type epidermal nuclei (*) reposition beneath the hyphae of P. medicaginis, surrounded by a large patch of ER. F, P. medicaginis hyphal contact triggers nuclear repositioning in the dmi3-1 mutant, but no ER accumulation. G and H, The contact with O. maius hyphae does not induce ER accumulation or nuclear repositioning in either wild-type or dmi3-1 roots. The alignment of an epidermal cell nucleus with a hypha was only observed occasionally, as shown in G (*). h, Hypha; hp, hyphopodium. Bars = 20 μm.
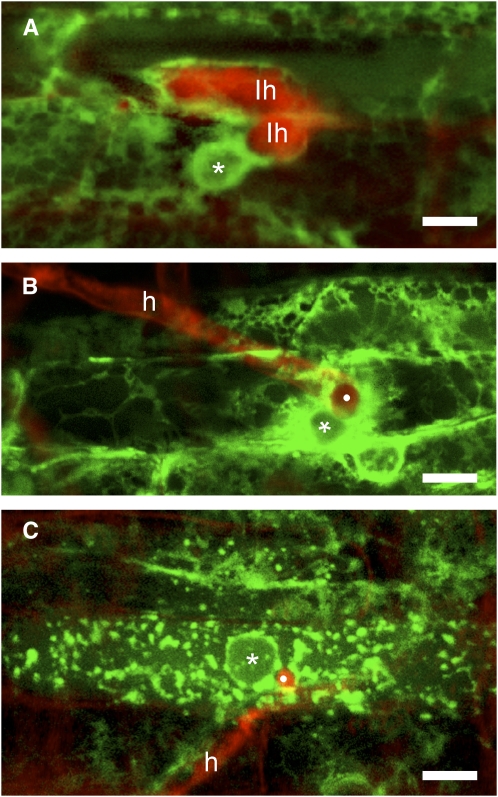
Figure 2.
Wild-type and dmi3-1 epidermal cell organization upon fungal entry. Identical fluorescent ER labeling and color coding are as in Figure 1. A, G. margarita intracellular hyphae (Ih) inside two adjacent epidermal cells, about 12 h after hyphopodium adhesion. The plant nucleus (*) of the lower cell is still in the vicinity of the transcellular tunnel hosting the fungus, but the dense ER accumulation characterizing the PPA has disappeared and loose ER cisternae surround the hyphae. B, About 48 h after inoculation with C. trifolii, an intracellular vesicle (white dot) is visible inside an epidermal cell from wild-type roots (see also Supplemental Fig. S1). The vesicle is surrounded by a dense accumulation of ER and in close vicinity to the nucleus (*). C, The entry of a C. trifolii hypha into the lumen of a dmi3-1 epidermal cell also occurs at about 48 hpi in the vicinity of the nucleus (*), but is associated with the disruption of the ER, which appears broken into clusters of roughly spherical bodies containing GFP. h, Hypha. Bars = 10 μm.
Epidermal cells of the dmi3-1 mutant only showed nuclear repositioning at fungus contact points but no sign of CA (Fig. 1B). Time-lapse observations did not reveal any further development of PPA-like responses or fungal penetration, as was also found with G. gigantea (Genre et al., 2005).
In conclusion, PPA-associated responses are DMI3 dependent, with the important exception of nuclear repositioning, which also occurs in the mutant.
CA in Response to C. trifolii Is DMI3 Dependent
A recent study has provided a description of root infection by a Colletotrichum species (Colletotrichum graminicola on maize [Zea mays]), demonstrating how this genus is not limited to aerial tissue colonization, but also colonizes the root system of its host plant (Sukno et al., 2008). In maize, the development of infectious structures mirrors the well-known stages of leaf infection, with the only exception that appressoria are never formed on the root epidermis, where they are functionally replaced by hyphopodia. We have applied a very similar protocol to that used by Sukno et al. (2008) to study the interaction between M. truncatula root organ cultures and C. trifolli race 2 in vitro. Although we have limited our observations to the early stages of the interaction (from inoculation to epidermal cell entry), the timing and developmental program of root infection was quite similar, within this time frame, to that described for C. graminicola in maize.
Wild-type epidermal cells responded within 24 hpi with a strong CA under the hyphopodium contact points, associated with nuclear repositioning (Fig. 1C). The ER patches that were observed were generally more extended than those triggered by AM fungi, with large interconnected cisternae. Fungal penetration, in the form of a short, roughly spherical hypha of about 10 μm in diameter (Supplemental Fig. S1A), developed at around 48 hpi, in agreement with the leaf infection process (Torregrosa et al., 2004). This occurred within the area of strong CA, in the vicinity of the nucleus (Fig. 2B) and among the GFP-labeled ER membranes. Later observations of the infected roots showed that areas of dead cells—revealed by the disappearance of GFP fluorescence—started to be visible at 72 to 96 hpi (Supplemental Fig. S2A).
The roots of the dmi3-1 mutant showed no signs of CA in the presence of C. trifolii hyphopodia, although nuclear repositioning under the contact area was clearly visible (Fig. 1D). The fungus penetrated the epidermal cells at around 48 hpi (Supplemental Fig. S1B), as observed in the wild type. At this stage, the ER of dmi3-1 epidermal cells broke down into clusters of small vesicles (Fig. 2C). GFP fluorescence then progressively and irreversibly disappeared and the nucleus became invisible in transmitted light images, suggesting the onset of epidermal cell death. Imaging of the later stages of infection (72–96 hpi) revealed narrow hyphae inside the lumen of the dead cells without detectable GFP fluorescence (Supplemental Fig. S2B).
In short, C. trifolii induced both nuclear repositioning and CA in wild-type M. truncatula and cell entry took place within the CA area. CA was instead absent in dmi3-1 roots, which only showed nuclear repositioning. Importantly, the mutation did not seem to affect overall fungal colonization, with the exception that epidermal cell death occurred at least 1 d earlier than for the wild type (Fig. 2C).
P. medicaginis Elicits CA Prior to Cell Death
P. medicaginis is known to infect roots and stems of M. truncatula with a necrotrophic strategy (Kosuta et al., 2003; O'Neill et al., 2003). Hyphae secrete toxins, including brefeldin, which rapidly kill the plant cells. In our experiments, hyphae were observed adhering to the root and following the surface of the epidermis. Fungal contact induced both nuclear repositioning and CA in wild-type epidermal cells (Fig. 1E). Wide areas of dead epidermal cells—where GFP fluorescence was no longer detectable—appeared around the contact areas 24 hpi (Supplemental Fig. S2C), confirming the necrotrophic development of the infection. Cells surrounding the dead area often showed a focal assembly of ER as well as nuclear repositioning, suggesting the spread of defense responses (Takemoto et al., 2003) to the neighboring tissue.
P. medicaginis contact induced nuclear repositioning, but no CA, in the epidermal cells of the dmi3-1 mutant (Fig. 1F). Areas of dead cells appeared at around 24 hpi, as was observed for the wild type (Supplemental Fig. S2D).
In brief, P. medicaginis triggered both nuclear repositioning and CA in the contacted wild-type cells, while the dmi3-1 mutation prevented formation of the CA, without apparently affecting fungal development.
Absence of Cellular Reorganization following Contact with O. maius
O. maius is an endomycorrhizal fungus that develops symbiotic interactions with the roots of Ericaceae, where it differentiates intracellular hyphal coils (Perotto et al., 1995). Unlike Gigaspora, this fungus is not an obligate biotroph and can also grow as a saprobe. Its interaction with M. truncatula was limited to mycelial growth on the root surface, where the thin hyphae (1.5 μm in diameter) did not develop any recognizable adhesion or penetration structure. Epidermal cells of wild-type M. truncatula did not show any visible response in terms of either nuclear repositioning or CA (Fig. 1G). When mycelial growth completely covered the root surface, areas of dead cells were observed, possibly as a consequence of the strong degradative enzymatic activity generated by this fungus on the plant cell wall (Martino et al., 2000). The exposure of dmi3-1 roots to O. maius resulted in the same absence of cellular responses (Fig. 1H).
In conclusion, O. maius did not induce the cellular dynamics described in the other interactions. Neither nuclear repositioning nor CA was triggered in the root epidermal cells by the presence of hyphal contact. This lack of response was observed for both wild-type and dmi3-1 roots and is probably correlated with the nonhost relationship between M. truncatula and O. maius (Bonfante et al., 1980).
Expression Profiles for Exp-Like and ACRE264 during Biotic Associations
Upon fungal contact, the Exp-like gene is known to be more highly expressed in the wild type as compared to the dmi3-1 mutant of M. truncatula, while the reverse is true for ACRE264 (Siciliano et al., 2007). Since Exp-like is hypothesized to be involved in cell wall loosening and ACRE264 in the elicitation of resistance mechanisms, which are presumed to be limited in compatible interactions, we wanted to examine whether this differential regulation was specific to the AM interaction—like the PPA—or a more generic plant response to fungal contact. To this aim, RNA was extracted from pools of short (1 cm) root segments contacted by each of the fungal strains used for the cellular investigations described above. Real-time RT-PCR analysis confirmed the previous results of Siciliano et al. (2007) for the M. truncatula/G. margarita combination and revealed unexpected responses for the remaining interactions (Table I).
Table I.
Relative transcript levels of Exp-like and ACRE264 genes in root segments contacted by different fungi compared to control roots
The values indicate relative gene transcript levels according to the 2−ΔΔCt method. The figures are the average values of 2−ΔΔCt, calculated for the two biological replicates. Asterisks indicate statistically significant differences in gene expression levels between wild-type and dmi3-1 backgrounds for each biological replicate.
| Fungus | Interaction |
Exp-Like
|
ACRE264
|
||
|---|---|---|---|---|---|
| Wild Type | dmi3-1 | Wild Type | dmi3-1 | ||
| G. margarita | AM symbiont | 2.1* | 1.3* | 2.4* | 5.0* |
| P. medicaginis | Necrotrophic pathogen | 2.6* | 1.4* | 2.5* | 4.0* |
| C. trifolii | Biotrophic pathogen | 4.3 | 4.2 | 3.2 | 3.3 |
| O. maius | Incompatible symbiont | 53.9* | 2.3* | 3.8* | 1.0* |
The regulation of both genes in response to the necrotrophic pathogen P. medicaginis was remarkably similar to that induced by G. margarita. Again in this case, Exp-like was in fact more strongly up-regulated in the wild type than in the dmi3-1 mutant, while ACRE264 had an opposite trend. These results, coupled with the cellular analyses, are in agreement with the susceptibility of wild-type Medicago to Phoma infection (Castell-Miller et al., 2007) and have provided direct evidence for the role of DMI3 in plant cell response during the early steps of the interaction, despite the fact that fungal development was not altered in the mutant background.
By contrast, during the interaction with C. trifolii, both sequences were up-regulated, confirming the induction of defense-related genes even in compatible pathogenic interactions of Medicago (Yang et al., 2008). The same expression pattern was conserved in the mutant, where the fungus developmental program and its entry into the plant cell were not affected by the mutation (Supplemental Fig. S1), although cell death occurred at an earlier stage of fungal entry. The fact that the dmi3-1 mutation affected CA but did not influence the Exp-like or ACRE264 expression, indicates that the regulation of these genes in this pathogenic interaction does not involve DMI3.
The noncompatible mycorrhizal fungus O. maius induced a very strong up-regulation of both genes in the wild-type roots, in spite of the absence of any evident cell response. This can be related to the plant cell wall degradation that this fungus can produce by means of its lytic enzymes (Martino et al., 2000), which in turn are expected to activate defense responses on the one hand (involving ACRE264) and cell wall remodeling on the other (involving Exp-like). In contrast, in the presence of the dmi3-1 mutation, the expression levels of both genes drop dramatically, suggesting that DMI3 activity also plays a role in interactions between mycorrhizal fungi and nonhost plants.
Physical Stimulation Triggers Nuclear Repositioning
To distinguish the effects produced by living fungi from those caused by simple physical contact, the epidermal cell surface was stimulated using a micromanipulator, equipped with a 10-μm-tipped microneedle. This action triggered a rapid movement of the nucleus to the contact area in wild-type roots that was completed within about 20 min (Fig. 3A), depending on the initial distance of the nucleus from the needle tip. No evidence of ER patch development was observed, even upon longer stimulation, suggesting that CA did not occur. Nuclear movement was abolished by a 0.5-μm latrunculin-B treatment (which also immobilized the ER), suggesting its dependency on the actin cytoskeleton (Supplemental Fig. S3A). Nuclear repositioning was not observed when a 1-μm-tipped microneedle was used (Supplemental Fig. S3B). When 10-μm needles were used to stimulate dmi3-1 mutants, the epidermal cells showed a rapid breakdown of the ER (Fig. 3B), with the initial appearance of clustered fluorescent vesicles, followed by the diffusion of GFP in the cell lumen and finally the complete disappearance of fluorescence within a few seconds (Fig. 3C). As in the wild type, 1-μm tips did not trigger any visible cell response (data not shown).
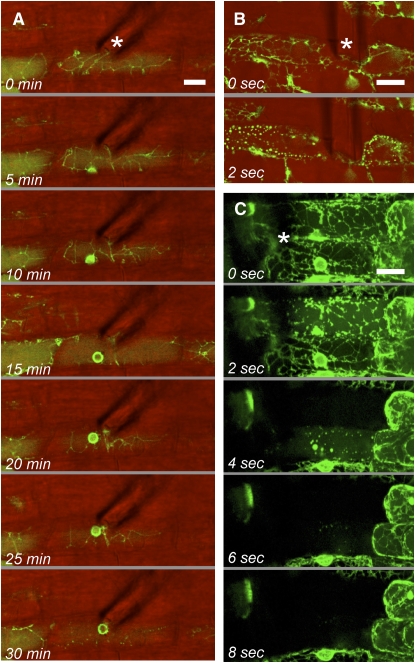
Figure 3.
Epidermal cell responses to physical stimulation with a microneedle (*) in wild-type (A) and dmi3-1 (B and C) roots. A, Microneedle contact triggers nuclear repositioning underneath the needle tip within 20 min. B, Physical stimulation of the dmi3-1 cells induces the rapid onset of ER disruption into clusters of roughly spherical fluorescent vesicles. C, Longer observations show the progressive break down of the ER, associated with the diffusion of GFP into the cytoplasm and eventually the complete disappearance of fluorescence within 8 s. No recovery of GFP fluorescence could be observed thereafter. A and B, Single optical sections (green) showing GFP fluorescence overlaid with bright-field images (red). C, Projections of optical sections showing only GFP fluorescence. Bars = 10 μm.
In conclusion, the cellular dynamics induced by the physical stimulation of wild-type root epidermal cells were limited to nuclear repositioning at the contact site with 10-μm-tipped needles, while the same stimulus triggered the rapid collapse of the ER membranes in dmi3-1 mutants.
DISCUSSION
Epidermal and cortical cells of mycorrhizal host plants anticipate fungal colonization by activating a process of cytoplasmic reorganization that has been termed the PPA (Genre et al., 2005, 2008). This finding raised the question about the specificity of such cell responses to AM fungi. By examining a range of different biotic and abiotic stimuli we have been able to identify both common elements (e.g. nuclear movement) as well as specific traits (e.g. PPA formation) among the different responses that were induced in host cells.
Cellular Responses
Nuclear repositioning appears to be a general, nonspecific response to physical stimulation. Regardless of what is touching the epidermal cell wall—symbiotic fungi, pathogenic fungi, or glass capillaries—the nucleus becomes positioned in close vicinity to the contact area, on condition that the touching object is larger than about 2 μm. The hyphopodia of G. margarita reach a diameter of over 15 μm, those of C. trifolii about 10 μm, while P. medicaginis hyphae have an average diameter of 4 μm. The only cases where nuclear repositioning was not observed were in response to contact with the thin hyphae (1.5 μm) of O. maius or 1-μm-tipped microneedles, suggesting that a size factor is involved. Nuclear repositioning has been observed in other experimental systems and has been described as traumatotactic movement (Pappelis et al., 1974; Goodbody and Lloyd, 1990). Such nuclear movements are believed to accelerate the cell response to an external stimulus. Within plant cells, the central vacuole represents an obstacle to rapid signal transduction between membrane-associated receptors and the nucleus. Positioning the nucleus closer to the stimulated area can lead to faster activation of the target genes (Gross et al., 1993), and can be an alternative (or a complement) to the intracellular transport of signaling endosomes (Geldner and Robatzek, 2008).
An important conclusion from our study is that nuclear movement can be uncoupled from CA, suggesting that different mechanisms mediate the displacement of different cell organelles. The cell dynamics described in analogous experiments performed on cotyledons of Arabidopsis (Arabidopsis thaliana; Hardham et al., 2008), where different cell components accumulate at the contact site, might suggest a species, tissue, or organ specificity in the response to physical stimuli.
CA, instead, correlates with the contact by compatible fungi, and was observed whenever a symbiont or a pathogen of M. truncatula was in contact with the surface of an epidermal cell. Three out of four tested fungi induced the accumulation of ER around the contact site. The only one that did not was O. maius, which is possibly not recognized as a root-interacting fungus by the nonhost plant. In this context it will be interesting to examine whether an AM fungus induces any response in a nonmycorrhizal plant, such as Arabidopsis. Both P. medicaginis and C. trifolii elicit the assembly of large patches of ER that may occupy the whole side of the cell in contact with the fungus. By contrast, G. margarita causes the accumulation of ER cisternae within a smaller area, more strictly localized to the contact site. This morphological difference between the CAs probably reflects the contrasting roles played by these cytoplasmic rearrangements: either acting as a first defense to block fungal attack by secreting cell wall appositions (O'Connell and Panstruga, 2006) or building a specific corridor to allow the symbiont entry (Bonfante and Genre, 2008). This interpretation could also provide a mechanistic explanation for the responses observed in the dmi3-1 mutant background: By hampering CA organization in the presence of a pathogen, the mutation of DMI3 compromises the cell's first defense line, allowing fungal development in spite of the elicitation of ACRE264. In the case of AM fungal contact, on the contrary, the lack of CA affects PPA development, thus resulting in the arrest of fungal development (Genre and Bonfante, 2007).
Experimental evidence suggests that the PPA is involved in the building of the apoplastic compartment where the intracellular hyphae of the AM fungus are hosted (Genre et al., 2008). For this reason, the assembly of a PPA is expected only when fungal growth across a living cell is required. Our hypothesis is confirmed by the lack of PPA assembly in the response to P. medicaginis, where live cell penetration does not occur. In the case of C. trifolii, fungal penetration occurs within the CA, but trans-cellular PPAs, associated with the second nuclear migration, are never detected. Haustorium formation within an area of CA that includes the nucleus has been described in Arabidopsis leaf epidermal cells attacked by Erysiphe cichoracearum (Koh et al., 2005). Nuclear movement away from the contact area was previously reported for the interaction with the biotroph Uromyces vignae (Heath et al., 1997), which also develops trans-cellular hyphae in living cells, but the description, based on differential interference contrast video microscopy, does not suggest the presence of a PPA-like structure.
In conclusion, an AM fungus does not only set off a specific type of CA in the contacted epidermal cell, but also triggers the assembly of a more complex transcellular apparatus, the PPA, devoted to fungal accommodation.
The Impact of the dmi3-1 Mutation on the Early Responses to Biotic and Abiotic Stimuli
DMI3 is a calcium and calmodulin-dependent protein kinase belonging to the so-called SYM genes (Parniske, 2008). These genes are essential for the establishment of both root nodulation and AM formation in legumes (Lévy et al., 2004; Parniske, 2008). Mutations in DMI3 affect root colonization in both symbioses by blocking epidermal penetration, and they also alter interaction with plant growth-promoting bacteria (Sanchez et al., 2005).
The induction of cell death, observed upon physical stimulation, is a striking feature of the dmi3-1 mutant. Nevertheless, a possible analogy can be found in observations of Esseling et al. (2004) who reported the touch sensitivity of mutants in DMI2, another member of the SYM genes. In those experiments, the contact of growing root hair tips with a microneedle rapidly stopped apical growth and cytoplasmic streaming. We therefore tested physical stimulation to epidermal cells of dmi2-2 root cultures expressing GFP-HDEL (kindly provided by M. Chabaud and D. Barker, INRA/CNRS, Castanet Tolosan, France). ER disruption was rapidly triggered by microneedle contact and proceeded with the same pattern and timing as described for dmi3-1 (data not shown). It is therefore likely that both mutations lead to a similar hypersensitivity to touch. These observations show that a block in the signaling pathways that are essential for both initiating nodulation and AM formation also affects other basic functions of the epidermal cell, such as the ability to respond to exogenous physical contact, an event that is likely to occur frequently in the soil. Accepting this conclusion implies either that thigmostimulation has a significant role upstream of the SYM signaling pathway, or that key proteins in the pathway have other roles in the perception of physical pressure on the cell wall (Haswell et al., 2008), thus leading to new questions about their involvement in a broader cell mechanism.
To date, the involvement of SYM genes in pathogenic plant/fungus interactions has only been investigated once: Mellersh and Parniske (2006) inoculated Uromyces loti onto leaves of Lotus japonicus carrying mutations in each of six different SYM genes (including the DMI3 ortholog), and reported a colonization process identical to that shown on wild-type plants. Our results, which focused on the root response, now provide additional important information. We have observed that CA, a cell response shared by the AM biotroph and by the two pathogens, is clearly DMI3 dependent, thus demonstrating the crucial role of this gene during the contact phase. Since, in the case of the AM biotroph, CA represents the initiation of the PPA, then DMI3 functionality is indispensable for live cell entry. On the other hand, it is dispensable for the necrotrophic phase of pathogenic infections. In the case of C. trifolii, cell death occurs upon cell entry, suggesting that the switch from biotrophy to necrotrophy is anticipated.
These data therefore suggest that DMI3 is also involved in the host cell response to pathogens, despite the fact that fungal development is not modified in the mutant background.
M. truncatula Responds to Fungal Contact by Activating the Exp-Like and ACRE264 Genes
Global transcriptomic approaches have recently been used to investigate whether plants respond in comparable ways to symbiotic and pathogenic fungi (Paszkowski, 2006). Such an analysis on rice (Oryza sativa) has revealed the presence of a common set of regulated genes among AM, hemibiotrophic, and necrotrophic fungal interactions (Guimil et al., 2005). Even though in a different context, and on a smaller scale, the two genes investigated here (Exp-like and ACRE264) are both up-regulated in all the interactions we have studied with the wild type. With the exception of C. trifolii, their expression pattern has been demonstrated to be DMI3 dependent.
ACRE264 was reported to be up-regulated in tobacco (Nicotiana tabacum) cell suspension cultures expressing the Cladosporium fulvum Cf-9 resistance gene, and identified as a protein kinase required for the resistance to fungal strains expressing the Avr9 gene (Rowland et al., 2005). The gene identified in M. truncatula shows similarity to this tobacco ACRE264 (Siciliano et al., 2007) and belongs to the group of genes that are up-regulated in the shoots of M. truncatula after infection of Glomus intraradices or treatment with Xanthomonas campestris (Liu et al., 2007). A related sequence of rice (identified as a putative Avr9 elicitor response protein) was also activated by both AM colonization and Magnaporte grisea infection (Guimil et al., 2005). These data support our observation of ACRE264 up-regulation in all the interactions of wild-type M. truncatula, although to a different extent (weaker activation by G. margarita and P. medicaginis, stronger activation in C. trifolii and O. maius). ACRE264 was also differently regulated in the dmi3-1 background, depending on the nature of the interaction. As expected, ACRE264 up-regulation in the presence of G. margarita, coupled with the absence of PPA, led to the colonization block, while the same up-regulation in the presence of the pathogens was apparently not sufficient to block fungal development. This is possibly due to the lack of the DMI3-dependent CA, which cancels one of the first defense mechanisms (O'Connell and Panstruga, 2006).
Expansins are a superfamily of proteins that play a crucial role in cell wall loosening (Sampedro and Cosgrove, 2005). Expansin-coding genes have been demonstrated to be up-regulated not only in AM (Journet et al., 2002; Liu et al., 2003; Weidmann et al., 2004; Siciliano et al., 2007), but also during nodulation (Giordano and Hirsch, 2004) and nematode attack (Gal et al., 2006; Wieczorek et al., 2006). Our data are largely in agreement with these results, suggesting that fungal contact and/or fungal enzymatic activity may stimulate plant cell wall plasticity, irrespective of the fungal nutritional strategy.
Medicago and Oidiodendron: Host versus Nonhost Responses
Our results on the O. maius interaction with M. truncatula fall into a different category, that of host versus nonhost plant/fungus combinations. As expected, O. maius never shows biotrophic development on M. truncatula (Bonfante et al., 1980). On the contrary, its thin hyphae grow over the roots as they would on organic debris. This fungus has the ability to grow as a saprobe and its enzymatic arsenal is rich in pectinases and other cell wall degrading enzymes (Martino et al., 2000). The lack of nuclear movement and CA are peculiar features of this nonhost interaction. We hypothesize that the lack of nuclear repositioning in the presence of O. maius contact is due to the absence of an appropriate physical stimulation to the epidermal cell, while the lack of CA could be partly related to this, or might reveal a more complex scenario including the misrecognition of O. maius as a potentially interacting microbe. The molecular dialogue between the plant and the fungus, which is well known in pathogenic interactions (O'Connell and Panstruga, 2006) and has also started to be deciphered in AM (Genre and Bonfante, 2007; Parniske, 2008), may well be missing or abnormal in this nonhost plant/fungus coupling.
Nonetheless, the saprotrophic growth of O. maius on M. truncatula roots most likely leads to plant cell wall degradation by the many lytic enzymes of the fungus. It will be interesting to investigate whether this activity is the stimulus that triggers Exp-like and ACRE264 regulation, even in the absence of visible cellular responses. Our cellular and molecular analyses suggest that we are probably observing the outcome of a diverse regulatory mechanism. Plant responses in a nonhost mycorrhizal interaction have not been investigated previously, but one can speculate that such interactions trigger similar plant basal defense mechanisms to those already described in Arabidopsis interacting with noncompatible pathogens (Lipka et al., 2005).
CONCLUSION
The comparison of the cell dynamics shown by M. truncatula roots, when challenged by an AM fungus, two pathogens, and an ericoid mycorrhizal fungus, has facilitated the dissection of some of the complex events that take place in an epidermal cell prior to fungal colonization. When the plant-fungus combination is compatible—irrespective of the beneficial or pathogenic nature of the interaction—the plant cell perceives fungal contact by activating a range of responses. The common responses can be interpreted as a general prealert status (Genre and Bonfante, 2007). Nuclear movement is a central event in these responses, and always precedes live cell penetration. Since similar preinfection dynamics have also been described following rhizobium contact (Fournier et al., 2008), we can suggest that a basic program of cell response to microbes is operating in M. truncatula roots, but it is finely modulated to give different outcomes depending on the nature of the interacting organism.
MATERIALS AND METHODS
Plant and Fungal Materials
Experiments were performed with Agrobacterium rhizogenes-transformed root cultures derived from both wild-type Medicago truncatula Jemalong A17 and dmi3-1 (TRV25; Sagan et al., 1998), kindly provided by M. Chabaud and D. Barker (INRA/CNRS, Castanet Tolosan, France). Root explants were grown on solid M medium (Chabaud et al., 2002) in vertically oriented petri dishes to stimulate their positive geotropism. Both root lines were expressing GFP-HDEL, a GFP tag that specifically labels the ER (Genre et al., 2005). This tag was chosen from the available constructs as the most suitable to monitor cell organization as it allows the simultaneous observation of nuclear and ER positions and provides general information on cytoplasm distribution.
The fungal strains used in this study were the AM symbiont Gigaspora margarita strain BEG 34, the hemibiotrophic pathogen Colletotrichum trifolii race 2 strain MUT 3930 (Richard O'Connell, Max Planck Institute, Cologne, Germany), the necrotrophic pathogen Phoma medicaginis var. medicaginis strain MUT 2049 (Mycotheca Universitatis Taurinensis, University of Torino, Italy), and the ericoid mycorrhizal fungus Oidiodendron maius strain Zn (MUT 1381).
Spores of G. margarita were produced using clover (Trifolium repens) cultures on sand, surface sterilized, and stored at 4°C according to Bécard and Fortin (1988). C. trifolii, P. medicaginis, and O. maius were grown on solid M medium in petri dishes at 25°C in the dark.
Conditions for in Vivo Microscopic Observation
The targeted inoculation technique developed by Chabaud et al. (2002) to study AM interactions and modified by Genre et al. (2005) was further adapted for the inoculation with the pathogenic fungi. Germinated spores of G. margarita were transferred to vertically oriented petri dishes containing a growing M. truncatula transgenic root explant, to obtain rapid root colonization. C. trifolii, P. medicaginis, and O. maius were inoculated as mycelial plugs placed in the vicinity of the roots. In all cases, the root and fungus were covered with 1 μL of sterile water, on top of which a thin (25 μm) gas-permeable film (bioFOLIE 25; Sartorius AG) was laid. This allowed continuous confocal microscopy observations and prevented medium desiccation or contamination, without perturbing the development of fungus-root interactions. None of the samples used for the micromanipulation experiments or gene expression analyses (see below) were covered with bioFOLIE to allow direct access to the root.
Micromanipulator-Mediated Abiotic Stimulation
A Leica Mechanical Micromanipulator was fixed to the microscope table and equipped with a single microneedle holder. The microneedles were prepared from borosilicate capillaries (WPI, 1.5 mm external diameter/0.84 internal diameter) using a Narishige PE-21 vertical capillary puller, according to the manufacturer's instructions, to obtain tips of the desired length and a tip diameter of about 1 μm. To obtain 10-μm tips, the microneedles were then gently filed with a forceps under a stereomicroscope. For thigmostimulation, microneedles with either a 1- or 10-μm tip diameter were placed in the holder, positioned in the vicinity of the root surface, and then gently moved vertically to place them in contact with the cell wall. This contact was revealed by a slight displacement of the sample out of the focal plane.
For the latrunculin-B treatments, 10-μm-tipped needles were plugged into a water-filled PVC tube (2 mm diameter, 15 cm long) connected to a micropipette, which provided the compensation pressure to load and unload the capillary with liquids. The effective expulsion of the needle load was verified by using a 200 μg/mL TRITC solution and observing the flow of fluorescent material (excitation light 543 nm; emission filter 580–620 nm) out of the needle tip under the confocal microscope (data not shown). To observe the effect of actin depolymerization on the microneedle-induced nuclear movement, a 0.5 μm latrunculin-B (Sigma) solution in sterile distilled water was loaded into the microneedle and expelled after cell wall contact by operating the micropipette plunger wheel.
Confocal Microscopy
Epidermal cell responses to fungal contact or abiotic stimulation were observed and followed in detail with a Leica TCS SP2 confocal microscope, using a long-distance 40× water-immersion objective (HCX Apo 0.80). The Ar laser band of 488 nm was used to excite the GFP and to collect transmitted light images of the samples. An emission window of 500 to 525 nm was used to image GFP fluorescence. The Leica Confocal Software application was used to recode the transmitted light channel in a negative gray scale, improve contrast, and finally false color it in red, before assembling the final pictures, where the GFP and transmitted light channels were overlaid. This improved the overall contrast of the images, where both the red hyphae and the green signal of the GFP clearly stand out on the dark background. The Leica Confocal Software application was also used for time-lapse imaging.
RNA Extraction and Analysis
Segments of inoculated roots were selected under a stereomicroscope to identify the sites of fungal contact with the root epidermis. Equivalent root segments were sampled from noninoculated control roots. All the samples were immediately frozen with liquid nitrogen and stored at −80°C. Two independent RNA extractions from 30 root segments were made for each experimental condition. RNA extraction was performed with the SV total RNA Isolation System kit (Promega). An additional precipitation was then performed overnight with an equal volume of LiCl 6 m. All the samples were quantified and checked through NanoDrop 1000 (Thermo Scientific) analysis. After extraction, a DNase treatment was performed with Turbo DNase (Ambion) according to the manufacturer's instructions and DNA contamination was checked using a one-step RT-PCR kit (Qiagen) with MtTefa (translation elongation factor 1alpha) primers (Vieweg et al., 2005).
Real-Time RT-PCR
For real-time reactions, cDNAs were obtained from 500 ng of extracted RNA using a High Capacity cDNA RT kit (Applied Biosystems), according to the manufacturer's indications. The cDNAs were then one-fifth diluted and tested by PCR with MtTefa primers. Real-time experiments were carried out in a final volume of 20 μL containing 10 μL of 2× Power Sybr green PCR master mix (Applied Biosystems), 0.2 mm primers, and 1 μL of cDNA. Real-time RT-PCR was performed on the StepOne Real-Time PCR system (Applied Biosystems) using the following thermocycler program: 10 min preincubation at 95°C, followed by 50 cycles of 15 s at 95°C, and 1 min at 60°C. Each amplification was followed by melting curve analysis 60°C to 94°C with a heating rate of 0.3°C every 15 s. Only specific products were detected in all the experiments. The 2−ΔΔCt method (Kenneth and Schmittgen, 2001) was utilized to calculate the relative expression levels and the data were analyzed using the Kruskal-Wallis test (P < 0.05) to verify the statistical significance of the observed differences. Target gene expression was normalized with the MtTefa gene. The primer pairs used were: MtTefa-f (AAGCTAGGAGGTATTGACAAG) and MtTefa-r (ACTGTGCAGTAGTACTTGGTG) for MtTefa (Vieweg et al., 2005); Exp-f (GTCGGTCTTATGGGGCAGTA) and Exp-r (ATTGCGAACCTTGACTCCAC) for Exp-like; and Acre-f (GGTTATGCGGCTCCTGAGTA) and Acre-r (CCATTCCACCAAGTTTTTCC) for ACRE264. The Exp-like and ACRE264 primer pairs were designed according to Siciliano et al. (2007).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EC366179 and EC366239.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Bright-field micrographs of wild-type (A) and dmi3-1 (B) roots stained with cotton blue to label C. trifolii hyphal walls.
Supplemental Figure S2. Late stages (about 96 hpi) of infection by C. trifolii (A and B) and P. medicaginis (C and D) in wild-type (A and C) and dmi3-1 (B and D) roots.
Supplemental Figure S3. Abolishment of nuclear repositioning in response to physical stimulation of the epidermal cell wall in wild-type roots.
Supplementary Material
Acknowledgments
We are grateful to R. O'Connell for useful discussions and for kindly providing C. trifolii; to M. Chabaud and D. Barker for kindly providing the GFP-expressing root lines; and to the MUT for kindly providing P. medicaginis. The San Paolo Company is acknowledged for contributing to the acquisition of a Leica confocal microscope for the Laboratory of Advanced Microscopy in Turin.
This work was supported by grants to P.B. from the Italian Ministry of Education (Progetti di Rilevanza Nazionale 2006), from the University of Turin (60%), Consiglio Nazionale delle Ricerche (Biodiversity Project), and the EU INTEGRAL Project. G.O. was supported by a Master and Back grant from the Regione Sardegna.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Paola Bonfante (p.bonfante@ipp.cnr.it).
The online version of this article contains Web-only data.
References
- Bécard G, Fortin A (1988) Early events of vesicular-arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol 108 211–218 [DOI] [PubMed] [Google Scholar]
- Bonfante P, Genre A (2008) Plants and arbuscular mycorrhizal fungi: an evolutionary-developmental perspective. Trends Plant Sci 13 492–498 [DOI] [PubMed] [Google Scholar]
- Bonfante P, Gianinazzi-Pearson V, Martinengo L (1980) Ultrastructural aspects of endomycorrhiza in the Ericaceae. IV. Comparison of infection by Pezizella ericae in host and non-host plants. New Phytol 98 329–333 [Google Scholar]
- Castell-Miller CV, Zeyen RJ, Samac DA (2007) Infection and development of Phoma medicaginis on moderately resistant and susceptible alfalfa genotypes. Can J Plant Pathol 29 290–298 [Google Scholar]
- Chabaud M, Venard C, Defaux-Petras A, Bécard G, Barker DG (2002) Targeted inoculation of Medicago truncatula in vitro root cultures reveals MtENOD11 expression during early stages of infection by arbuscular mycorrhizal fungi. New Phytol 156 265–273 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD (2001) Plant pathogens and integrated defense responses to infection. Nature 411 826–833 [DOI] [PubMed] [Google Scholar]
- Esseling JJ, Lhuissier FG, Emons AM (2004) A nonsymbiotic root hair tip growth phenotype in NORK-mutated legumes: implications for nodulation factor-induced signaling and formation of a multifaceted root hair pocket for bacteria. Plant Cell 16 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier J, Timmers ACJ, Sieberer BJ, Jauneau A, Chabaud M, Barker DG (2008) Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol 148 1985–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal TZ, Aussenberg ER, Burdman S, Kapulnik Y, Koltai H (2006) Expression of a plant expansin is involved in the establishment of root knot nematode parasitism in tomato. Planta 224 155–162 [DOI] [PubMed] [Google Scholar]
- Geldner N, Robatzek S (2008) Plant receptors go endosomal: a moving view on signal transduction. Plant Physiol 147 1565–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Bonfante P (2007) Check-in procedures for plant cell entry by biotrophic microbes. Mol Plant Microbe Interact 9 1023–1030 [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Faccio A, Barker DG, Bonfante P (2008) Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG (2005) Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17 3489–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano W, Hirsch AM (2004) The expression of MaEXP1, a Melilotus alba expansin gene, is upregulated during the sweetclover-Sinorhizobium meliloti interaction. Mol Plant Microbe Interact 17 613–622 [DOI] [PubMed] [Google Scholar]
- Goodbody K, Lloyd CW (1990) Actin filaments line-up across Tradescantia epidermal cells, anticipating wound-induced division planes. Protoplasma 157 92–101 [Google Scholar]
- Gross P, Julius C, Schmelzer E, Hahlbrock K (1993) Translocation of cytoplasm and nucleus to fungal penetration sites is associated with depolymerization of microtubules and defense gene activation in infected, cultured parsley cells. EMBO J 12 1735–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimil S, Chang H, Zhu T, Sesma A, Osbourn A, Roux C, Ioannidis V, Oakeley EJ, Docquier M, Descombes P, et al (2005) Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci USA 102 8066–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gus-Mayer S, Naton B, Hahlbrock K, Schmelzer E (1998) Local mechanical stimulation induces components of the pathogen defense response in parsley. Proc Natl Acad Sci USA 95 8398–8403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardham AR, Jones DA, Takemoto D (2007) Cytoskeleton and cell wall function in penetration resistance. Curr Opin Plant Biol 10 342–348 [DOI] [PubMed] [Google Scholar]
- Hardham AR, Takemoto D, White RG (2008) Rapid and dynamic subcellular reorganization following mechanical stimulation of Arabidopsis epidermal cells mimics responses to fungal and oomycete attack. BMC Plant Biol 8 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ (2005) Signaling in the arbuscular mycorrhizal symbiosis. Annu Rev Microbiol 59 19–42 [DOI] [PubMed] [Google Scholar]
- Haswell ES, Peyronnet R, Barbier-Brygoo H, Meyerowitz EM, Frachisse J (2008) Two MscS homologs provide mechanosensitive channel activities in the Arabidopsis root. Curr Biol 18 730–734 [DOI] [PubMed] [Google Scholar]
- Heath MC, Nimchuk ZL, Xu H (1997) Plant nuclear migrations as indicators of critical interactions between resistant or susceptible cowpea epidermal cells and invasion hyphae of the cowpea rust fungus. New Phytol 135 689–700 [Google Scholar]
- Jaffe MJ, Leopold AC, Staples RC (2002) Thigmo responses in plants and fungi. Am J Bot 89 375–382 [DOI] [PubMed] [Google Scholar]
- Journet EP, van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer M, Niebel A, Schiex T, Jaillon O, Chatagnier O, et al (2002) Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30 5579–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenneth JL, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Koh S, Andre A, Edwards H, Ehrhardt D, Somerville S (2005) Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J 44 516–529 [DOI] [PubMed] [Google Scholar]
- Kosuta S, Chabaud M, Lougnon G, Gough C, Dénarie J, Barker DG, Bécard G (2003) A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol 131 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Bednarek P, Schultze-Lefert P (2008) Secretory pathways in plant immune responses. Plant Physiol 147 1575–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané J, Lauber E, Bisseling T, et al (2004) A putative Ca2+ and calmodulin-dependant protein kinase required for bacterial and fungal symbioses. Science 303 1361–1364 [DOI] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Lantag J, Brandt W, Rosahl S, Scheel D, et al (2005) Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310 1180–1183 [DOI] [PubMed] [Google Scholar]
- Liu J, Blaylock LA, Endre G, Cho J, Town CD, VandenBosch KA, Harrison MJ (2003) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Maldonado-Mendoza I, Lopez-Meyer M, Cheung F, Town CD, Harrison MJ (2007) Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J 50 529–544 [DOI] [PubMed] [Google Scholar]
- Martino E, Coisson JD, Lacourt I, Favaron F, Bonfante P, Perotto S (2000) Influence of heavy metals on production and activity of pectinolytic enzymes in ericoid mycorrhizal fungi. Mycol Res 104 825–833 [Google Scholar]
- Mellersh D, Parniske M (2006) Common symbiosis genes of Lotus japonicus are not required for intracellular accommodation of the rust fungus Uromyces loti. New Phytol 170 641–644 [DOI] [PubMed] [Google Scholar]
- O'Connell RJ, Panstruga R (2006) Tête-à-tête inside a plant cell: establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytol 171 699–718 [DOI] [PubMed] [Google Scholar]
- O'Neill NR, Bauchan GR, Samac DA (2003) Reactions in the annual Medicago spp. core germ plasm collection to Phoma medicaginis. Plant Dis 87 557–562 [DOI] [PubMed] [Google Scholar]
- Pappelis AJ, Pappelis GA, Kulfinski FB (1974) Nuclear orientation in onion epidermal cells in relation to wounding and infection. Phytopathology 64 1010–1012 [Google Scholar]
- Parniske M (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6 763–775 [DOI] [PubMed] [Google Scholar]
- Paszkowski U (2006) Mutualism and parasitism: the yin and yang of plant symbioses. Curr Opin Plant Biol 9 364–370 [DOI] [PubMed] [Google Scholar]
- Perotto S, Peretto R, Faccio A, Schubert A, Varma A, Bonfante P (1995) Ericoid mycorrhyzal fungi: cellular and molecular bases of their interactions with the host plant. Can J Bot 73 S557–S568 [Google Scholar]
- Rowland O, Ludwig AA, Merrick CJ, Baillieul F, Tracy FE, Durrant WE, Fritz-Laylin L, Nekrasov V, Sjölander K, Yoshioka H, et al (2005) Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9-dependent disease resistance in tomato. Plant Cell 17 295–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan M, de Larambergue H, Morandi D (1998) Genetic analysis of symbiosis mutants in Medicago truncatula. In C Elmerich, A Kondorosi, WE Newton, eds, Biological Nitrogen Fixation for the 21st Century, Vol 31. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 317–318
- Sampedro J, Cosgrove DJ (2005) The expansin superfamily. Genome Biol 6 242–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez L, Weidmann S, Arnould C, Bernard AR, Gianinazzi S, Gianinazzi-Pearson V (2005) Pseudomonas fluorescens and Glomus mosseae trigger DMI3-dependent activation of genes related to a signal transduction pathway in roots of Medicago truncatula. Plant Physiol 139 1065–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano V, Genre A, Balestrini R, Cappellazzo G, DeWitt P, Bonfante P (2007) Transcriptome analysis of arbuscular mycorrhizal roots during development of the prepenetration apparatus. Plant Physiol 144 1455–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukno SA, Garcia VM, Shaw BD, Thon MR (2008) Root infection and systemic colonization of maize by Colletotrichum graminicola. Appl Environ Microbiol 4 823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D, Hardham AR (2004) The cytoskeleton as a regulator and target of biotic interactions in plants. Plant Physiol 136 3864–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D, Jones DA, Hardham AR (2003) GFP-tagging of cell components reveals the dynamics of subcellular re-organization in response to infection of Arabidopsis by oomycete pathogens. Plant J 33 775–792 [DOI] [PubMed] [Google Scholar]
- Torregrosa C, Cluzet S, Fournier J, Huguet T, Gamas P, Prosperi JM, Esquerré-Thugayé M, Dumas B, Jaquet C (2004) Cytological, genetic, and molecular analysis to characterize compatible and incompatible interactions between Medicago truncatula and Colletotrichum trifolii. Mol Plant Microbe Interact 17 909–920 [DOI] [PubMed] [Google Scholar]
- Vieweg MF, Hohnjec N, Küster H (2005) Two genes encoding different truncated hemoglobins are regulated during root nodule and arbuscular mycorrhiza symbioses of Medicago truncatula. Planta 220 757–766 [DOI] [PubMed] [Google Scholar]
- Wieczorek K, Golecki B, Gerdes L, Heinen P, Szakasits D, Durachko DM, Cosgrove DJ, Kreil DP, Puzio PS, Bohlman H, et al (2006) Expansins are involved in the formation of nematode-induced syncytia in roots of Arabidopsis thaliana. Plant J 48 98–112 [DOI] [PubMed] [Google Scholar]
- Weidmann S, Sanchez-Calderon L, Descombin J, Chatagnier O, Gianinazzi S, Gianinazzi-Pearson V (2004) Fungal elicitation of signal transduction related plant genes precedes mycorrhiza establishment and requires the DMI3 gene in Medicago truncatula. Mol Plant Microbe Interact 17 1385–1393 [DOI] [PubMed] [Google Scholar]
- Yang SM, Gao MQ, Xu CW, Gao J, Deshpande S, Lin S, Roe BA, Zhu H (2008) Alfalfa benefits from Medicago truncatula: the RCT1 gene from M. truncatula confers broad-spectrum resistance to anthracnose in alfalfa. Proc Natl Acad Sci USA 105 12164–12169 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.