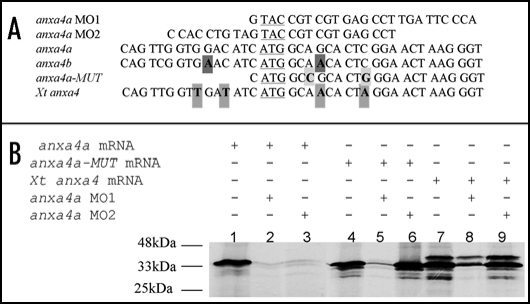
Figure 6.
The anxa4 MO2 knocks down anxa4a but not Xt anxa4 translation. (A) Alignment of the 5′ sequences of X. laevis wild type and mutant anxa4 cDNA and its X. tropicalis homolog showing the complementarity of anxa4a MO1 and MO2 in relation to these cDNAs. The ATG is underlined and mismatch residues are indicated in bold. Four nucleotides (in grey) differ between Xt anxa4 cDNA and the MO2 sequence. Two mismatch nucleotides created in anx4a-MUT cDNA are highlighted in light grey. Two nucleotides (in dark grey) are different between anxa4b and MO2 sequences. (B) anxa4a MO2 does not interfere with Xt anxa4 translation; autoradiograph of an SDS-PAGE analysis gel of in vitro translated 35S-Methionine radio-labelled anxa4 proteins. 0.5 µg of anxa4a mRNA was incubated alone (lane 1) or in combination with 10 µg of anxa4a MO1 (lane 2) or MO2 (lane 3). Both MO block the translation of anxa4a mRNA. 0.5 µg of anxa4a-MUT mRNA was incubated either alone (lane 4) or in combination with 10 µg of anxa4a MO1 (lane 5) or MO2 (lane 6). Translation of the mutant mRNA is severely affected by MO1 but unaffected by MO2. 0.5 µg of Xt anxa4 mRNA was incubated alone (lane 7) or in combination with 10 µg of anxa4a MO1 (lane 8) or MO2 (lane 9). MO2 does not block the translation of Xt anxa4 whereas the addition of MO1 affects its translation. These results demonstrate that Xt anxa4 can be used to rescue MO2.