Figure 4.
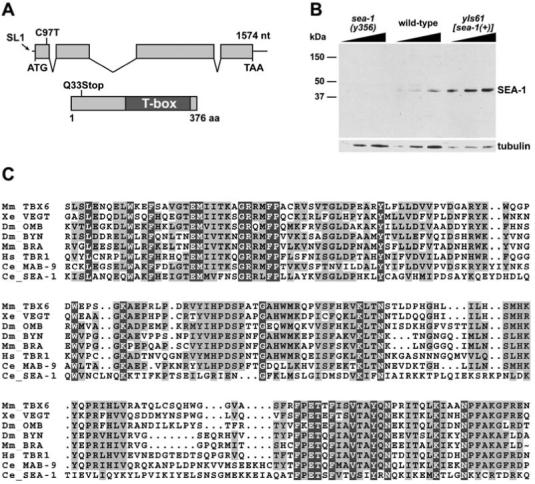
sea-1 Encodes a T-Box Protein
(A) The predicted genomic and protein structures of sea-1 are diagrammed. The site to which the SL1 trans-spliced RNA leader is attached to the sea-1 mRNA is represented on the DNA by an arrow. sea-1(y356) is a C to T transition at nucleotide 97 after the likely translational start site, replacing a glutamine codon with an ochre stop codon at position 33. SEA-1 contains a T-box DNA binding domain in the C-terminal half.
(B) Western blot of wild-type, sea-1 mutant, and sea-1(+) overexpressing embryonic extracts probed with anti-SEA-1 antibodies, with a tubulin loading control. SEA-1 protein is not detectable in sea-1(y356) mutant embryo extract but is present in wild-type extract and overexpressed in yIs61[sea-1(+)] extract. The predicted size of wild-type SEA-1 is 42.7 kDa.
(C) Alignment of SEA-1 T-box domain with T-box domains from other species. SEA-1 is significantly diverged from other T-box proteins. The SEA-1 T-box domain shares 27% identity with that of mouse BRACHYURY and frog VEGT. In contrast, the more conventional T-box domain of the C. elegans MAB-9 protein shares 52% identity with both BRACHYURY and VEGT. Similarly, the T-box domains of BRACHYURY and VEGT are 52% identical.
