Abstract
Hepatitis B virus (HBV) infection is a major health concern with more than two billion individuals currently infected worldwide. Despite the prevalence of infection, gaining a complete understanding of the molecular mechanisms of HBV infection has been difficult because HBV cannot infect common immortalized cell lines. HepG2.2.15, however, is a well established version of the HepG2 cell line that constitutively expresses HBV. Therefore, comparative proteomics analysis of HepG2.2.15 and HepG2 may provide valuable clues for understanding the HBV virus life cycle. In this study, two-dimensional blue native/SDS-PAGE was utilized to characterize different multiprotein complexes from whole cell lysates between HepG2.2.15 and HepG2. These results demonstrate that two unique protein complexes existed in HepG2.2.15 cells. When these complexes were excised from the gel and subjected to the second dimension separation and the proteins were sequenced by mass spectrometry, 20 non-redundant proteins were identified. Of these proteins, almost 20% corresponded to heat shock proteins, including HSP60, HSP70, and HSP90. Antibody-based supershift assays were used to verify the validity of the distinct protein complexes. Co-immunoprecipitation assays confirmed that HSP60, HSP70, and HSP90 proteins physically interacted in HepG2.2.15 but not HepG2 cells. We further demonstrated that down-regulation of HSP70 or HSP90 by small interfering RNA significantly inhibited HBV viral production but did not influence cellular proliferation or apoptosis. Consistent with these results, a significant reduction in HepG2.2.15 HBV secretion was observed when the HSP90 inhibitor 17-allylamino-17-demethoxygeldanamycin was used to treat HepG2.2.15 cells. Collectively these results suggest that the interaction of HSP90 with HSP70/HSP60 contributes to the HBV life cycle by forming a multichaperone machine that may constitute therapeutic targets for HBV-associated diseases.
Hepatitis B virus (HBV)1 is a member of the hepadnavirus family. HBV infection is associated with transient and chronic liver inflammation (1), and it has been well documented that long term chronic HBV infection can result in liver cirrhosis and hepatocellular carcinoma. Currently more than two billion people are estimated to be infected by HBV worldwide, and more than 350 million people are believed to be chronically infected. Chronically infected patients are at a greater risk (∼100-fold) of developing hepatocellular carcinoma (2).
During the past 2 decades, many principles of HBV infection have been resolved. Specifically the infectious viral genome has been cloned, the viral proteins have been well characterized, and the mechanism of viral DNA replication has been uncovered (3). In addition, the in vivo course of viral infection has been characterized with the help of closely related viruses like duck and woodchuck HBV (3). Despite this progress, a number of stages in the HBV life cycle, including the mechanisms of viral entry, uncoating, assembly, delivery, and secretion, remain to be fully elucidated.
The human HBV virion cannot directly infect common immortalized cell lines. Therefore, the lack of an efficient in vitro cell culture system in which HBV is propagated has long impeded the study of the viral life cycle (4). HepG2.2.15 is a well established hepatoblastoma cell line derived from HepG2 cells that constitutively expresses HBV as a consequence of the integration of a 2-fold version of the HBV genome. HepG2.2.15 cells support full replication of HBV and secrete hepatitis B surface antigen (HBsAg), hepatitis B e antigen (HBeAg), and virions into the culture medium (5). Chimpanzees intravenously inoculated with the HepG2.2.15 culture medium have been reported to develop typical hepatitis (6). Collectively these data indicate that the HepG2.2.15 cell line could represent an ideal model for the analysis of host-virus interactions. However, despite the fact that the HepG2.2.15 cell line has existed for 20 years, our knowledge of these cells, especially compared with the parental cell line (HepG2), is limited. Therefore, comparative biological analysis between HepG2.2.15 and HepG2 is expected to provide insight necessary for elucidating the HBV life cycle.
It has been suggested that nearly all biological and biochemical processes are performed by protein complexes (7). This is particularly true in viral infections where host and/or virus proteins assemble into complexes necessary to perform required processes in viral entry, replication, assembly, trafficking, and secretion (8). Therefore we propose that utilizing HepG2 and HepG2.2.15 cells to identify and characterize multiprotein complexes unique to HepG2.2.15 cells constitutes a unique methodology by which to elucidate the network of protein-protein interactions that regulate viral protein functions and HBV infection. In utilizing this methodology, the limiting factor for identifying protein complexes is the method for protein separation. Recently developed blue native (BN)-PAGE offers an attractive proteomics solution for identification of functional protein complexes.
BN-PAGE permits a high resolution separation of multiprotein complexes under native conditions (9). This technique consists of polyacrylamide gel electrophoresis where the non-denaturing compound Coomassie Blue G-250 is added to both the sample and to the electrophoresis buffers to confer a negative charge on the protein complexes so they can migrate intact toward the anode (9, 10). Using this methodology, many samples can be concurrently separated during a single electrophoretic run, and a direct comparison of protein complexes readily allows for the identification of disorders and direct further functional analysis. In combination with second dimension SDS-PAGE, a BN gel strip can be excised and placed horizontally on the SDS-PAGE to screen a series of proteins in the complex (11). This 2D method (2D BN/SDS-PAGE) has been successfully utilized to identify changes in protein complex composition from tissue homogenates, cells, and cell fractions during the analysis of physiological states and to diagnose pathological disorders such as Parkinson disease and cytochrome c oxidase deficiency (12–14). To date, protein complex composition and dynamics between different samples have only been analyzed following BN-PAGE and 2D SDS-PAGE. In the current study, an optimized strategy was used where distinct complexes were first identified following BN-PAGE, and the band for each unique complex was excised from the gel and subjected to the second dimension SDS-PAGE. We believe that this protocol might not only alleviate labor but also has the potential to enhance the resolution of the protein complex separation by reducing the cross-ratio of noise to signal in a gel.
In the present study, the optimized 2D BN/SDS-PAGE strategy was used to visualize the “footprints” of protein complexes between HepG2.2.15 and HepG2 cells. A total of 35 protein bands from two distinct complex bands in HepG2.2.15 cells were observed and analyzed by mass spectrometry. As a result of this analysis, 20 proteins were identified that corresponded to almost 20% of cellular heat shock proteins (HSPs) and a significant number of proteins that are part of enzyme complexes. Because previous studies have shown that HSPs are implicated in viral replication, we further investigated whether HSPs chaperone machinery is involved in the HBV life cycle in HepG2.2.15 cells. We believe that as result of this study we have identified and characterized a number of functional protein complexes that will provide a methodology for the analysis of endogenous multiprotein complexes between different cell types and cells exposed to different conditions (i.e. stimulated and unstimulated, normal and tumor, and wild type and knock-out).
EXPERIMENTAL PROCEDURES
Cell Culture—
HepG2 and HepG2.2.15 were grown in minimal Eagle's medium (Hyclone) supplemented with 10% fetal calf serum (Invitrogen), 100 units/ml penicillin/streptomycin, and 2% l-glutamine at 37 °C and 5% CO2 in a humidified atmosphere. HepG2.2.15 cells were additionally maintained in medium containing 380 μg/ml G418. The medium was changed every 2–3 days.
BN-PAGE and Antibody Supershift Assays—
Cells (107) were lysed in 500 μl of BN solution buffer 25BTH20G (25 mm BisTris-HCl, 20% glycerol, pH 7.0) supplemented with 2% dodecyl maltoside and protease inhibitor mixture (Roche Applied Science). After incubation on ice for 40 min, the lysates were centrifuged at 15,000 × g at 4 °C for 30 min. The supernatant containing protein complexes was removed and stored on ice. The protein concentration was determined by reducing agent compatible and detergent compatible (RC DC) assay (Bio-Rad).
BN-PAGE and 2D BN/SDS-PAGE were performed as described previously (9, 10). For the blue native first dimension, 80 μg of protein was added to BN sample buffer (1× BisTrisACA (100 mm BisTris-HCl, 500 mm 6-aminocaproic acid, pH 7.0), 30% glycerol, 5% Coomassie Brilliant Blue G-250) and loaded onto the gel. BN-PAGE was performed in a Hoefer™ SE250 unit (GE Healthcare) using a 4% stacking gel and a 5–13.5% separating gel. GE Healthcare HMW Native protein markers were used as the molecular weight standard. The cathode buffer (50 mm Tricine, 15 mm BisTris) containing 0.01% (w/v) Coomassie Brilliant Blue G-250 and the anode buffer (50 mm BisTris-HCl, pH 7.0) were chilled to 4 °C prior to use. Samples were separated overnight at 10 °C.
For the antibody-based gel supershift assays, approximate 80 μg of HepG2.2.15 cellular protein extracts were combined with 1 μg of anti-HSP60 antibody (Santa Cruz Biotechnology) and incubated on ice for 30 min before loading the mixture on the BN-PAGE. An equivalent concentration of HepG2.2.15 cellular protein lysates was combined with 1 μg of an irrespective antibody (normal IgG; Santa Cruz Biotechnology) used as a negative control.
2D BN/SDS-PAGE—
The dye front was allowed to run off of the gel, and the differentiated protein complex bands were then cut out from the first dimension of the BN-PAGE and equilibrated for 30 min in 1× SDS loading buffer at room temperature. The extracted bands were rinsed with deionized water and then placed on top of a separation gel consisting of a 12% Laemmli SDS gel with a 5% upper gel and sealed with 1% hot agarose solution. The second dimension run was performed according to standard protocols (6, 8–10). Gels were stained with Coomassie Brilliant Blue (10% (v/v) acetic acid, 40% (v/v) methanol, 0.25% (w/v) Coomassie R-250).
Identification of Proteins by Mass Spectrometry—
Protein spots from 2D BN/SDS-PAGE were excised and cut into small particles manually. After in-gel digestion with trypsin, the extracted peptide mixtures were loaded onto a nanoscale LC-ESI-Q-TOF MS instrument (Q-TOF Micromass spectrometer, Waters) for protein identification as described elsewhere (15). Briefly nanoscale reverse phase HPLC (PepMap C18, LC Packings) of the peptide mixture was carried out on a CapLC liquid chromatography system including three pumps, A, B, and C (Waters). The flow rate was set at 2.5 μl/min and split into approximately 0.2 μl/min prior to the precolumn and analytical column. Samples were injected at a flow rate of 20 μl/min with pump C by the autosampler, and salts and other impurities were removed on the precolumn of 300-μm inner diameter × 5-mm PepMap C18, 3-mm spherical particles with pore diameter of 10 nm (LC Packings). The precolumn was connected in the 10-port switching valve and switched to the analytical column after the sample was desalted. The separation was performed by running a nonlinear gradient: 4% B (water/ACN (20:80, v/v) with 0.1% formic acid) in 0.1–10 min for injection; 4–50% B in 10–40 min, 50–95% B in 40–49 min, 95% B in 49–54 min, and 95–4% B in 54–58 min. After 7-min equilibration with mobile phase A (water/ACN (98:2, v/v) with 0.1% formic acid), another analysis could be run. The CapLC was coupled on line with a Q-TOF Micromass spectrometer (Waters) for detection and protein identification.
For the analysis of peptide mixture by CapLC-ESI MS/MS, lyophilized peptide mixtures were solubilized with 15 μl of mobile phase A and loaded by autosampler attached to the CapLC system onto a trapping column (PepMap C18, LC Packings). Peptides were then directly eluted from the capillary C18 column into the mass spectrometer at about 200 nl/min for acquiring tandem MS data. The instrument was calibrated with a multipoint calibration using Glu-fibrinopeptide B (1570.6774 Da; Applied Biosystems), and residual errors of all less than 3 mDa were assured. The positive ion mode was used, the spray voltage was set at 3.2 kV, and the spray temperature was 80 °C. Collision energy was set at 30 V for MS/MS, and the voltage of the microchannel plate was set at 2850 V. After the acquisition of full-scan mass spectra, four MS/MS channels were chosen to acquire mass spectrometric data from the four most intense ions in series every 1.1 s for seven times using dynamic exclusion within 55 s. MS/MS data were processed using MassLynx version 4.0, and the resulting MS/MS data sets were exported into the pkl files.
Protein Identification Criteria—
Mascot search engine (version 1.9, Matrix Science) was used to search all of the tandem mass spectra. The NCBInr human and virus protein databases (NCBInr 20060211) were used for the search and were restricted to tryptic peptides. Cysteine carbamidomethylation and methionine oxidation were selected as variable modifications. One missing cleavage was allowed. Precursor error tolerance was set to <0.3 Da, and MS/MS fragment error tolerance was set to <0.2 Da. All the proteins identified should have at least two peptides matched and individual ions scores greater than 35 with expect value <0.05. One-peptide hits were manually analyzed to ascertain the accuracy of protein identification. In the manual analysis, the criterion used for a true identification is that the masses of all the major peaks (typically more than seven peaks) in a MS/MS spectrum had to match those of the theoretically calculated fragment ions.
Western Blotting and Immunoprecipitation—
Western blotting and immunoprecipitation procedures were described previously (43). Anti-HSP60, anti-HSP70, anti-HSP90, anti-GAPDH, and anti-β-actin antibodies were purchased from Santa Cruz Biotechnology. Cells were lysed in Nondenaturing Lysis Buffer (Applygen) supplemented with protease inhibitor mixture (Roche Applied Science). For Western blot analysis, protein concentrations were determined as discussed above, equivalent concentrations of protein samples were boiled in electrophoresis sample buffer, and proteins were resolved by 12% SDS-PAGE. After transferring to nitrocellulose membranes, filters were blocked for 1 h in blocking buffer (20 mm Tris-HCl, pH 7.5, 137 mm NaCl, 0.05% (v/v) Tween 20 (TBST) containing 5% dried milk) and then incubated with the primary antibodies diluted in blocking buffer. The membrane was washed with TBST and then incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h prior to visualization of the bands by an enhanced chemoluminescence system (Pierce). For immunoprecipitation experiments, lysates were precleared and then incubated (on a rotator) with an antibody against HSP60, HSP70, or HSP90 at 4 °C for 4 h or overnight. Following incubation, the immune complexes were precipitated with protein A/G-agarose (Santa Cruz Biotechnology) for 2 h at 4 °C, the samples were washed, proteins were separated by SDS-PAGE, and expression was determined by immunoblot analysis.
siRNA and Transfection—
The four pairs of 19-nt siRNA duplexes targeting HSP70 and HSP90 corresponding to the respective coding regions were designed and synthesized by GenePharma. HSP70 siRNA-1 was raised against the sequence corresponding to nt 363–382 (5′-GACCAAAGCTGATCTCATA-3′), HSP70 siRNA-2 corresponded to nt 792–811 (5′-GAAAGAAGAGGAAGATAAA-3′), HSP70 siRNA-3 corresponded to nt 1343–1362 (5′-CAGAAGACAAGGAGAATTA-3′), HSP70 siRNA-4 corresponded to nt 1439–1458 (5′-CTGAGCTGCTGCGCTATCA-3′), HSP90 siRNA-1 corresponded to nt 437–456 (5′-CTTCTATGGTTCTGACAAA), HSP90 siRNA-2 corresponded to nt 952–971 (5′-GACTTCTATACCTCCATTA-3′), HSP90 siRNA-3 corresponded to nt 1038–1057 (5′-CCTTCGAGATGCCAAACTA-3′), and HSP90 siRNA-4 corresponded to nt 1845–1864 (5′-GGAAGAATTTGAACATCAA-3′). All siRNA sequences were compared against human sequences deposited in GenBank™ databases with BLAST (Basic Local Alignment Search Tool) searches using the NCBI (National Center for Biotechnology Information) tools.
For siRNA experiments, HepG2.2.15 cells were grown overnight in 12-well plates. Cells were transfected with 100 pmol of siRNA using Lipofectamine™ 2000 (Invitrogen) as instructed by the manufacturer. Forty-eight hours after transfection, protein extracts were analyzed by immunoblot analysis to confirm protein knockdown.
ELISA and Quantitative PCR—
The levels of HBsAg and HBeAg in culture media from cells, respectively, were determined by ELISA according to the manufacturer's instructions. Viral DNA was isolated from cell culture media using the High Pure Viral Nucleic Acid kit (Roche Applied Science). Prior to isolation of the viral DNA, traces of non-encapsidated DNA were removed by DNase treatment (30 min at 37 °C) of the culture media with 4 units/ml DNase I (final concentration; New England Biolabs). Viral DNA was quantitated by real time PCR in an AbiPrism4000 (PerkinElmer Life Sciences) as described previously (16). Each ELISA and real time PCR experiment was performed four times in triplicate.
Cell Proliferation and Apoptosis Assays—
HepG2.2.15 cells were plated in triplicate in 24-well plates at a density of 105 cells/well and transfected with siRNA specific to HSP90 or HSP70 or with a nonspecific control siRNA. After transfection for 48 h, cell proliferation was analyzed by a methyl thiazolyl tetrazolium (MTT)-based assay. For each MTT assay, the medium in each well was replaced with 400 μl of medium containing MTT at 0.5 μg/μl, and plates were incubated at standard culture conditions. After 4 h of incubation, the MTT-containing medium was removed, 400 μl of DMSO was added to each well, and the plate was agitated for 10 min in the dark to dissolve the MTT-formazan crystals. Sample absorbance was recorded at 570 nm. Caspase-3 assays were performed to measure levels of apoptosis using a caspase-3 assay colorimetric kit (Kegene) as instructed by the manufacturer. The above experiments were performed in triplicate, and the results are presented as the mean ± S.D.
RESULTS
2D BN/SDS-PAGE Reveals That Two Distinct Multiprotein Complexes Containing HSP Chaperones Specially Exist in HepG2.2.15—
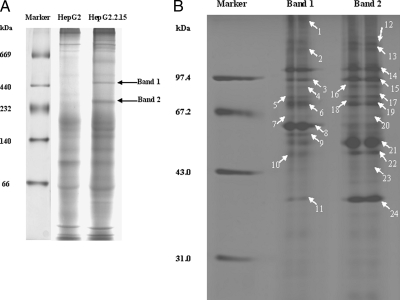
HepG2.2.15 cells are genetically derived from HepG2 and serve as an in vitro HBV production system. Therefore, to investigate differences in cellular protein complex formation in the presence of viral proteins, we utilized the BN-PAGE system. This technique was initially established for the analysis of mitochondrial protein complexes and more recently has been utilized to analyze the multiprotein complexes of whole cellular lysates that collectively have validated this technique as a method for a range of functional proteomics studies (9, 11). Here we utilized the BN-PAGE methodology to investigate differences in endogenous protein complexes formed in HepG2 and HepG2.2.15 cells. It was expected that utilizing this strategy would enable us to identify host proteins involved in the HBV life cycle. By examining whole cell lysates of HepG2 and HepG2.2.15 by BN PAGE, we identified two obvious protein complex bands that corresponded to a molecular mass of ∼450 and 250 kDa (Band 1 and Band 2). These reproducible bands are easily detectable and are uniquely expressed in HepG2.2.15 cells (see Fig. 1).
Fig. 1.
The difference of protein complexes between HepG2 and HepG2.2.15 cells. A, one-dimensional BN-PAGE proteomic map of whole cell lysates from HepG2 and HepG2.2.15 cells. Whole cell lysates were subjected to BN-PAGE, and the molecular mass markers are shown on the left. The gel was stained by Coomassie Brilliant Blue. Arrows represent the two distinct protein complexes that specially existed in HepG2.2.15 cells (Band 1 and Band 2). B, 2D BN/SDS-PAGE analysis of the two distinct protein complexes. Band 1 and Band 2 from the gel were excised, equilibrated, and separated by SDS-PAGE. Arrows represent successfully identified protein bands, and the corresponding numbers are shown in Table I.
To identify the individual proteins in each complex, the two distinct bands were excised from the gel, equilibrated, and horizontally subjected to 12% SDS-PAGE. It was determined by Coomassie Brilliant Blue staining that these protein complexes constituted 35 individual proteins (Fig. 1B). To identify each protein, each spot was excised, subjected to trypsin digestion, and analyzed by mass spectrometry. As a consequence of this analysis, 24 proteins (marked by arrows in Fig. 1B) were identified of which 20 were non-redundant proteins (Table I; matched peptide information is described in supplemental Table 1).
Table I.
Proteins identified from BN/SDS-PAGE
| Spota | Accession numberb | Protein name | Abbreviation | Theoretical molecular mass | Score MS/MSc | Matched peptides | Localization |
|---|---|---|---|---|---|---|---|
| Da | |||||||
| 1 | gi 3282771 | Actin-binding protein 278 | ABP-278 | 278,018 | 169 | 5 | Cytoskeleton |
| 2 | gi 4504743 | Integrin α2 | ITAG2 | 129,214 | 58 | 2 | Cytoskeleton |
| 3 | gi 306891 | 90-kDa heat shock protein | HSP90 | 83,242 | 140 | 7 | Cytoplasm |
| 4 | gi 21361368 | Pyrroline-5-carboxylate synthetase isoform 1 | ALDH18A1 | 87,248 | 67 | 2 | Mitochondrion |
| 5 | gi 49457432 | G22P1 | Ku70 | 69,829 | 73 | 3 | Cytoplasm, nucleus |
| 6 | gi 5729877 | Heat shock protein 70 | HSP70 | 70,854 | 49 | 2 | Cytoplasm |
| 7 | gi 88567 | Ribophorin II | RNP2 | 69,259 | 55 | 1 | Endoplasmic reticulum |
| 8 | gi 45595681 | Heat shock protein 60 | HSP60 | 61,016 | 397 | 35 | Mitochondrion, cytoplasm |
| 9 | gi 32015 | α-Tubulin | α-Tubulin | 49,761 | 72 | 2 | Cytoskeleton |
| 10 | gi 45044327 | Hydroxyacyl dehydrogenase | HADH | 51,262 | 76 | 2 | Mitochondrion |
| 11 | gi 31645 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 36,031 | 73 | 2 | Cytoplasm |
| 12 | gi 5729887 | IQ motif-containing GTPase-activating protein 2 | IQGAP2 | 180,500 | 212 | 6 | Cytoplasm, cytoskeleton |
| 13 | gi 31621305 | Leucine-rich PPRd motif-containing protein | LRP130 | 157,805 | 131 | 4 | Mitochondrion, nucleus |
| 14 | gi 62088648 | Heat shock protein gp96 | HSP gp96 | 92,411 | 221 | 6 | Endoplasmic reticulum |
| 15 | gi 306891 | Heat shock protein 90 | HSP90 | 83,242 | 418 | 12 | Cytoplasm, nucleus |
| 16 | gi 35038 | ATP-dependent DNA helicase II | Ku86 | 82,652 | 82 | 3 | Nucleus, cytoplasm |
| 17 | gi 1143492 | BiP | Bip | 72,071 | 262 | 8 | Cytoplasm |
| 18 | gi 49457432 | G22P1 | Ku70 | 69,829 | 251 | 12 | Cytoplasm, nucleus |
| 19 | gi 5729877 | Heat shock protein 70 | HSP70 | 70,854 | 102 | 4 | Cytoplasm, nucleus |
| 20 | gi 3273228 | Acyl-coenzyme A dehydrogenase | ACAD | 70,391 | 111 | 3 | Mitochondrion |
| 21 | gi 20151194 | Glutamate dehydrogenase | GLUD | 55,973 | 349 | 11 | Mitochondrion |
| 22 | gi 23468343 | Translation elongation factor 1 | EF1 | 50,109 | 155 | 4 | Cytoplasm |
| 23 | gi 33150754 | GTP-binding protein | CDC42 | 44,715 | 43 | 1 | Cytoplasm |
| 24 | gi 31645 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 36,031 | 52 | 1 | Cytoplasm |
The letters correspond to the marked letters on the gel in Fig. 1B. Spots 1–11 come from the 450-kDa complex, and spots 12–24 come from the 250-kDa complex.
NCBInr database accession number.
Score from Mascot (Matrix Science). Individual scores >35 indicate identity (p < 0.05).
Pentatricopeptide repeat.
The most intensely stained protein spot that was isolated from the 450-kDa complex on the two-dimensional gel (Spot 8) was shown to have an electrophoretic mobility of 60 kDa. This protein was identified as HSP60, a molecular chaperone that has been shown to be associated with HBV replication by interacting with and activating HBV polymerase (HBV pol) (17). Furthermore two additional spots of 90 and 70 kDa (Spots 15 and 19) were identified as HSP90 and HSP70. These proteins have been reported to cooperate in a chaperone complex that alters the conformation of substrate proteins involved in signal transduction and viral replication (18–20). An additional seven proteins were identified in the 450-kDa complex including four enzymes, two cytoskeletal proteins, and a DNA-binding protein. HSP90 and HSP70 also exist in the 250-kDa complex with a higher score (408 and 102, respectively). In addition to HSP70 and HSP90, 12 other proteins were identified in the 250-kDa complex. In our attempt to deduce their mutually potential association utilizing the Protein-Protein Interaction Database (Human Protein Reference Database), we determined that, remarkably, many of the protein interactions that were found in the 250-kDa complex were previously identified in vivo and/or in vitro (supplemental Table 2). In contrast, proteins identified in the 450-kDa complex did not show any evidence of a previously identified association with the noted exception of HSP70 and HSP90. Therefore we investigated that potential contribution the identified protein had on HBV production.
Validation of the 450-kDa Complex by Anti-HSP60 Antibody Supershift Assays—
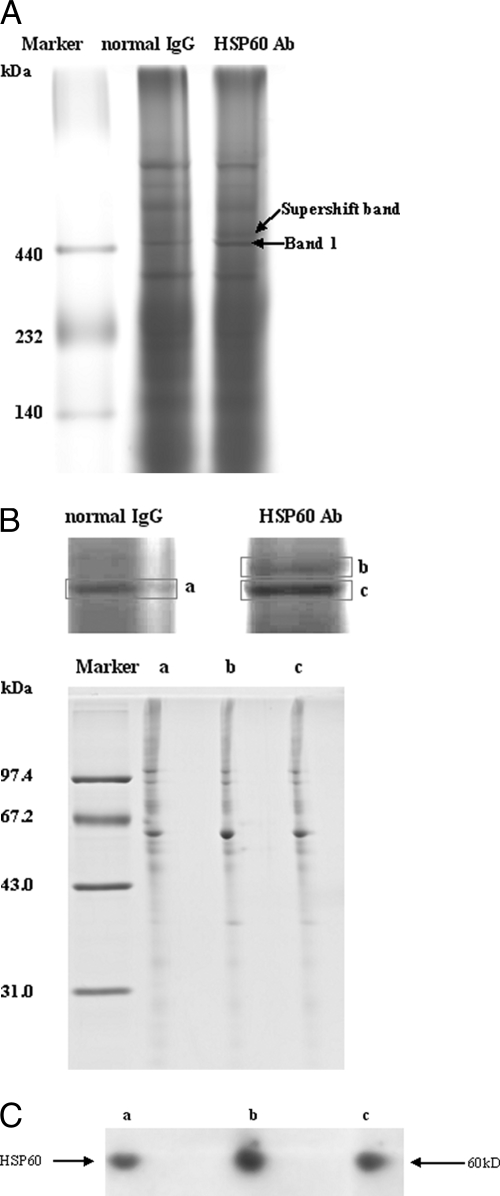
It was determined, as discussed above, that no obvious association existed between the proteins present in the 450-kDa complex. To verify the validity of this complex, we performed a supershift assay with an antibody to HSP60, which is the most abundant component of the 450-kDa complex. The whole cell lysates of HepG2.2.15 were incubated with anti-HSP60 antibody or normal IgG (negative control) prior to the BN-PAGE. It was expected that specific antibody binding would either retard migration of the HSP60-containing protein complex or interact with the protein and prevent it from entering the gel. Preincubation of cell lysates with an anti-HSP60 antibody resulted in the presence of a slower migrating complex as compared with the primary band (Band 1) but did not affect the mobility of Band 2 (Fig. 2A). Although the amount of the supershifted protein complex was relatively small, it was quite specific because normal IgG failed to produce a supershifted band. To further ascertain that the supershift band really originated from Band 1, gel strips corresponding to Band 1 in the lane of HepG2.2.15 cell lysates mixed with normal IgG (a), Band 1 (b), and supershift band (c) in the lane of HepG2.2.15 cell lysates mixed with anti-HSP60 antibody, respectively, were cut out and analyzed by 2D BN/SDS-PAGE. The protein maps of these three gel strips were identical (Fig. 2B). Western blot analysis using anti-HSP60 antibody also indicated that these three bands were comprised of HSP60 (Fig. 2C). These results suggested that HSP60 exclusively associates with the 450-kDa complex and confirmed the validity of this complex.
Fig. 2.
Verification of the validity of the distinct protein complexes that existed in 2.2.15 cells by BN-PAGE supershift assays. A, HepG2.2.15 whole cellular lysates were incubated with HSP60 antibody (Ab) and resolved by BN-PAGE. A supershift band based on Band 1 was formed (indicated by arrow); no supershift was observed when lysates were incubated with normal IgG. B, Band 1 in HepG2.2.15 cell lysates mixed with normal IgG (a), supershift band (b) mixed with anti-HSP60 antibody, and Band 1 (c) were excised and analyzed by BN 2D/SDS-PAGE. C, Western blot analysis confirmed that HSP60 existed in all three bands. The three bands were subjected to BN 2D/SDS-PAGE, transferred to nitrocellulose membrane, and immunoblotted for HSP60.
HSP60 Specially Associates with HSP70 and HSP90 in HepG2.2.15 Cells, Not in HepG2 Cells—
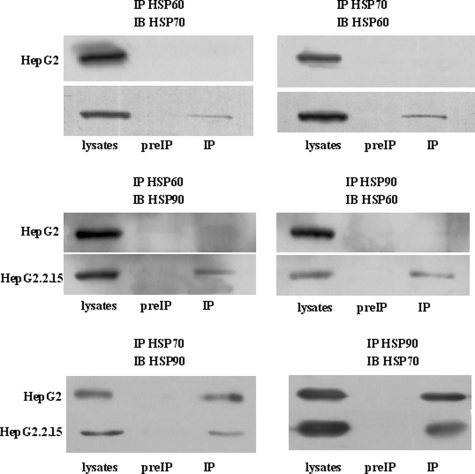
HSPs are commonly used to maintain protein quality during times of cellular stress (17). These proteins participate in protein complex formation, contribute to protein folding and extension, are essential for the assembly of polycomplexes, and function in protein transport between cell organelles among other essential functions. Furthermore HSPs have been implicated in the replication of several viruses with diverse genome structures and replication strategies (17–21). Therefore, it is not unexpected that a number of HSPs are present in the two identified protein complexes. As noted above, HSP60 is the most abundant component in one of complexes, and both HSP70 and HSP90 are consistently present in both complexes. More importantly, HSP60 and HSP90 have been reported to assist in HBV replication and assembly (17, 21). These data collectively led us to investigate whether HSP60, HSP70, and HSP90 might physically associate together to contribute to HBV production. Although our previously discussed studies failed to identify any HBV viral protein, we performed a series of coimmunoprecipitation assays to investigate this hypothesis. We first investigated whether HSP60, HSP70, and HSP90 would coimmunoprecipitate in HepG2.2.15 or HepG2 cellular lysates. Whereas no interaction was detected in HepG2 cellular lysates (Fig. 3, upper right and middle right), an interaction between these proteins was detected in HepG2.2.15 lysates following immunoprecipitation with an anti-HSP60 antibody (Fig. 3, upper left and middle left). Likewise HSP60 coimmunoprecipitated with HSP70 or HSP90 when antibodies specific to these proteins were used to probe HepG2.2.15 cellular lysates (Fig. 3, upper right and middle right). Furthermore HSP70 and HSP90 could be coimmunoprecipitated from either cellular lysate when either an HSP70 or HSP90 antibody was utilized. Taken together, these results indicate that HSP60, HSP70, and HSP90 form an HSP chaperone complex only in HepG2.2.15 cells. Although the conclusions that can be drawn from coimmunoprecipitation experiments are limited, these experiments can be a valuable starting point for studying contributions of HSP chaperone machinery to HBV production. Finally these studies validate the accuracy of the multiprotein complexes detected by BN-PAGE.
Fig. 3.
HSP60, HSP70, and HSP90 proteins physically associate in HepG2.2.15 cells, not in HepG2 cells. Upper panel, HepG2 and HepG2.215 cells were immunoprecipitated (IP) with HSP60 or HSP70 antibodies. Immunocomplexes were subjected to SDS-PAGE and immunoblot (IB) analysis for HSP70 (left) or HSP60 (right) expression. Middle panel, cells were immunoprecipitated with HSP60 or HSP90 antibodies prior to immunoblot analysis for HSP90 (left) or HSP60 (right). Lower panel, cell lysates were immunoprecipitated with anti-HSP90 or anti-HSP70 antibodies, and immunoblot analysis was performed for HSP70 (left) or HSP90 (right) expression. Corresponding IgG used as a negative control (preIP).
siRNA-mediated Down-regulation of HSP70 or HSP90 Inhibits HBV Production—
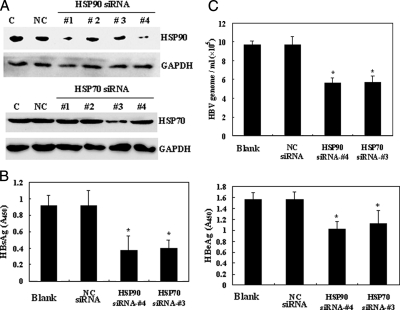
To determine whether HSP70 and HSP90 are necessary to regulate the HBV life cycle, four pairs of siRNA duplexes were designed for down-regulation of HSP70 and HSP90. The ability of the siRNA duplexes to repress HSP70 or HSP90 protein expression was confirmed by immunoblot analysis. An irrelevant siRNA was used as negative control for transfection, and GAPDH was used as the internal standard for Western blotting. Introduction of HSP70 siRNA-3 or the concurrent expression of HSP90 siRNA-1 and HSP90 siRNA-4 in HepG2.2.15 cells resulted in a significant decrease of HSP70 and HSP90 protein levels (Fig. 4A); other siRNA had no effect on the levels of these proteins. As a result of these preliminary studies, HSP70 siRNA-3 and HSP90 siRNA-4 were determined to be the most efficient at reducing HSP70 and HSP90 levels and were, therefore, utilized in the following experiments.
Fig. 4.
Inhibition of HBV virus production of HepG2.2.15 by siRNA-mediated down-regulation of HSP70 or HSP90. A, Western blot analysis validating HSP70 or HSP90 knockdown by siRNA. Negative control (NC) siRNA was used as the irrelevant control; GAPDH was used as the internal standard for sample loading. HSP70 siRNA-3 and HSP90 siRNA-4 were demonstrated to have the greatest effect on protein levels. Lane C, control. B, effects of down-regulation of HSP90 or HSP70 on extracellular encapsidated HBV secretion. Equivalent volumes of media from cells transfected with HSP70 siRNA, HSP90 siRNA, or negative control siRNA were analyzed by real time PCR for HBV genome copy numbers. C, cleared media from transfected cells were semiquantitated by ELISA for HBsAg (left) or HBeAg (right). Each ELISA and real time PCR experiment was performed in triplicate and repeated four times. The data shown are the mean ± S.D. of triplicate determinations. *, p < 0.05.
We first examined the effects of decreased HSP70 or HSP90 protein expression on HBV antigen secretion (HBsAg and HBeAg) in HepG2.2.15 cells. Cell culture supernatants were semiquantitatively analyzed by ELISA for HBeAg and HBsAg. A significant decrease in HBeAg and HBsAg secretion was detected in the supernatant of cells treated with HSP70 or HSP90 siRNA as compared with cells transfected with a control siRNA (Fig. 4B). Concurrently we examined the effects of HSP70 and HSP90 siRNA on the production of HBV viral particles by quantifying the presence of extracellular HBV genomes using real time PCR (Fig. 4C). It was determined that 48 h after transfection HepG2.2.15 cells treated with HSP70 or HSP90 siRNA demonstrated a significant decrease in the number of HBV genomes expressed. Collectively these data confirm that siRNA-mediated down-regulation of HSP70 or HSP90 inhibits HBV production in HepG2.2.15 cells.
Down-regulation of HSP70 or HSP90 Does Not Affect Cell Viability—
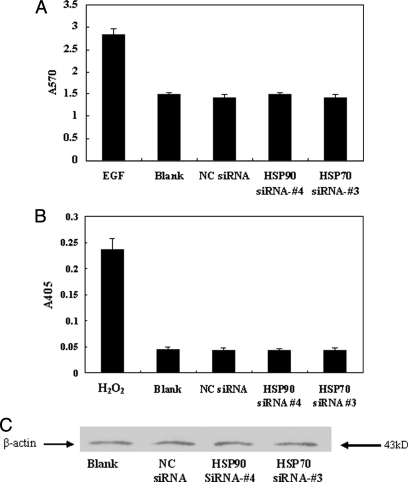
In normal eukaryotic cells, HSP70 and HSP90 are highly conserved and are essential stress proteins. Therefore, we next investigated whether down-regulation of either protein affected cell viability and, as a consequence, may affect viral production. To determine whether siRNA-mediated knockdown of HSP70 or HSP90 affected cell viability, an MTT assay was performed to investigate cellular proliferation, and caspase-3 assays were performed to measure levels of cellular apoptosis. We determined that siRNA-mediated knockdown of HSP70 or HSP90 had no significant effect on cellular proliferation as determined by MTT assay measured at A570 (Fig. 5A). Furthermore no increased induction of apoptosis was observed by caspase-3 assay (A405) in HepG2.2.15 cells treated with either HSP70 or HSP90 siRNA (Fig. 5B). A β-actin Western blot was used as an internal control to verify that equivalent numbers of cells were used in each assay (Fig. 5C). These results indicate that siRNA-mediated knockdown of neither HSP70 nor HSP90 affects cell viability.
Fig. 5.
Effects of HSP70 or HSP90 down-regulation on cell proliferation and apoptosis. HSP70, HSP90, or negative control (NC) siRNA-transfected HepG2.2.15 cells were analyzed for cellular proliferation by MTT assay (A) using 30 mg/liter epidermal growth factor (EGF)-treated sample as a positive control, apoptosis by caspase-3 assay (B) using 400 μm H2O2-treated sample as a positive control, and cell count as determined by Western blot analysis (anti-β-actin) (C). Data for all experiments are shown as the mean ± S.D. of three independent experiments. *, p < 0.05.
Inhibition of HSP90 by 17-Allylamino-17-demethoxygeldanamycin (17-AAG) Also Suppresses HBV Production—
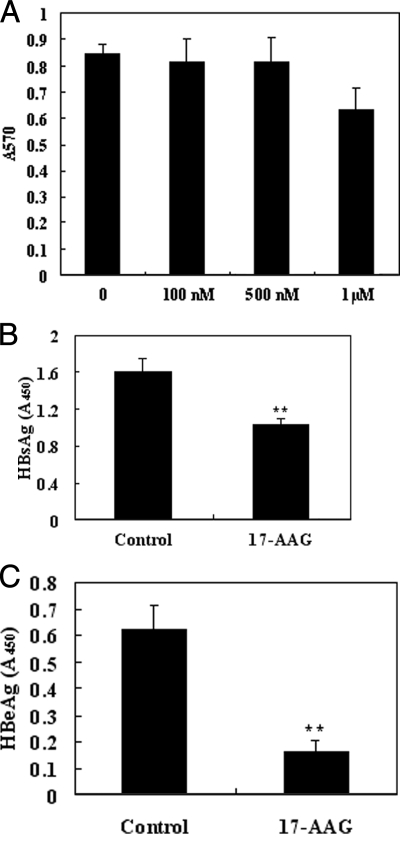
The cellular activities of HSP90 have been intensely studied, and it has been determined that HSP90 plays an integral role in protein homeostasis as well as serving as a selective chaperone of key signaling proteins in cancer survival and proliferation pathways. Therefore, HSP90 represents an attractive target for the development of a small molecule inhibitor. 17-AAG is currently one of the most studied small molecule inhibitors directed against HSP90 and is now the subject of multiple cancer clinical trials. Because we have demonstrated that down-regulation of HSP90 results in the suppression of HBV production, we next investigated whether 17-AAG might represent a promising therapeutic for the treatment of HBV-associated diseases. To test this hypothesis, HepG2.2.15 cells were treated for 24 h with 17-AAG using a range of doses (100 nm, 500 nm, and 1 μm). At the conclusion of the 24-h treatment, ELISA was used to quantify HBeAg and HBsAg expression levels in culture media. Simultaneously cell viability was determined by the MTT assays. The results of these experiments demonstrated that a 24-h regimen of HepG2.2.15 cells with a 500 nm dose of 17-AAG resulted in a significant reduction of extracellular HBsAg and HBeAg secretion (Fig. 6, B and C) but did not significantly affect cell viability (Fig. 6A).
Fig. 6.
HepG2.2.15 cells treated with HSP90 inhibitor (17-AAG) show reduced extracellular virus production. A, the dose effect of 17-AAG on HepG2.2.15 cell viability was investigated by MTT assay. Twenty-four hours after 17-AAG treatment, media from cells semiquantitatively were analyzed by ELISA for HBsAg (B) and HBeAg (C) levels. The assay was performed in triplicate, and the data are presented as the mean ± S.D. **, p < 0.01.
DISCUSSION
BN-PAGE was initially developed for the analysis of intact and functional mitochondrial membrane proteins and complexes; this technique offers the ability to separate native protein complexes without dissociating them (9). Consistent with its latter use, this technique has been successfully utilized for the isolation of protein complexes from biological membranes and total cell and tissue homogenates, to diagnose mitochondrial disorders, to determine the stoichiometry of a multiprotein complex, to identify physiological protein-protein interactions, and to investigate many other questions (11–14). In the current study, we utilized BN-PAGE to analyze differences in protein complexes present in HepG2 and HepG2.2.15 cell lysates to identify cellular proteins involved in the HBV life cycle.
When we examined the protein complexes that form in HepG2.2.15 cells, we identified two unique complexes that existed in these cells but not in the parental HepG2 cells and that corresponded to molecular masses of 250 and 450 kDa (Fig. 1A), respectively. Further examination of these complexes resulted in the identification of 20 unique cellular proteins that may be involved in viral infection and can be classified into four functional categories: heat shock proteins, proteases, DNA/RNA-binding proteins, and cytoskeletons.
Heat shock proteins are directly involved in protein biogenesis and contribute to various stages for this process beginning with synthesis of nascent chains through the assembly of multimeric complexes and have, therefore, been termed molecular chaperones (22). HSPs are also known to be involved in the life cycle of a number of viruses (17, 18, 20). Here we report the identification of HSP60, HSP70, and HSP90 from the protein complex found in Band 1 of HepG2.2.15 cells. In addition, a number of other cellular proteins were identified in Band 1; the role each protein may have on HBV viral activity is discussed.
HSP60 is predominantly localized to the mitochondria; however, it is also found in the cytosol, plasma membrane, and endoplasmic reticulum (23–25). HSP60 has been reported to assist in the correct folding of proteins and stabilize unfolded, labile proteins. Furthermore it was determined that HSP60 interacts with human HBV pol protein and that this interaction is necessary for the proper maturation of human HBV pol into the active state (19). Antisense oligodeoxynucleotides specifically directed against HSP60 resulted in decreased in HSP60 expression and severely reduced levels of replication-competent HBV without influencing cellular proliferation or capsid assembly (26).
HSP90 has also been reported to be an essential host factor for duck HBV and hepatitis C virus replication (20, 27). Previous studies have shown that HSP90 interacts with the viral reverse transcriptase thereby facilitating the formation of a ribonucleoprotein (RNP) complex between the polymerase and an RNA ligand. The RNP complex is required early in the replication process to mediate viral assembly and initiate DNA synthesis through a protein-priming mechanism (28). Several additional components of the HSP90 complex, including HSP70, HSP40, and p23, which are required for HSP90 function, are also key components of the polymerase complex and are required for reconstitution of viral reverse transcriptase activity (27, 28).
A number of additional cellular proteins were also identified in this complex. GAPDH is a key enzyme involved in glycolysis and gluconeogenesis as well as regulating protein kinase activity. HBV core particles derived from infected hepatocytes possess an associated kinase activity that phosphorylates hepatitis B core antigen, and the nucleocapsid may acquire sequential functions through selective phosphorylation. Because the associated kinase activity of the core protein may be derived from GAPDH, the potential exists that the phosphorylation of the core protein is involved in HBV RNA encapsidation (29, 30). LRP130 is a lectin-binding protein overexpressed in HepG2 cells that has an additional function as an RNA-binding protein that plays an integral role in nuclear RNP complexes associated with mature mRNA. Additional studies have determined that LRP130 interacts with IQGAP2, a prominent regulator of the cytoskeleton (31, 32). The third major protein identified in this complex was the Ku antigen (Ku70/Ku80), an abundant nuclear protein with multiple functions. Ku70/Ku80 is the regulatory subunit of DNA-dependent protein kinase, which regulates DNA replication and gene transcription through specific DNA sequences. Consistent with our findings that Ku70/Ku80 is associated with HBV proteins, a number of previous studies have demonstrated that Ku70 and Ku80 interacted with purified retroviral replication intermediates (33–35). Another key protein identified in our study was human ABP, which is a ubiquitous dimeric phosphoprotein that self-associates in non-muscle cells. A number of studies have suggested that ABP-278 may interact with HBV core protein to regulate genome packaging, DNA replication, and virion morphogenesis of HBV (36). Tubulins, which are microtubule monomers involved in intracellular movement of pathogens, were also identified in our analysis as interacting proteins. Microtubules and associated molecular motors are important for cytoplasmic transport and also for the establishment and stability of the bacterial replicative niche (37). In vitro, tubulin can stimulate vesicular stomatitis virus transcription, which suggested that tubulin was involved with the function of virus RNA polymerase (38). Collectively our data have demonstrated that a significant number of proteins identified in our analysis may be key components of cellular machinery utilized for viral infection and replication. In Band 1, a number of proteins were identified that could interact with HBV viral proteins including ABP-278, GAPDH, HSP60, HSP90, and HSP70. Therefore, we believe that during the course of the HBV life cycle proteins that exist in Band 1 may be involved in the formation of the HBV RNP complex.
Proteins identified in Band 2 were determined to be predominantly comprised of DNA/RNA-binding proteins that have been reported to be involved in the life cycles of other viruses including BiP, which is required for human cytomegalovirus virion assembly (39), and IQGAP, which interacts with Moloney murine leukemia virus matrix protein (40). During certain pathological conditions, including viral infection, the subcellular localization of some proteins may be altered to inhibit their cellular function or induce a specialized activity. Consistent with this idea, previous studies have demonstrated that during a hepatitis C infection, hepatitis C virus core protein causes the nuclear protein DDX3 to relocalize to the cytoplasm (41). Therefore, additional studies will be required to investigate the role that cellular proteins identified in this complex play in the HBV life cycle.
Coimmunoprecipitation experiments have demonstrated that HSP60, HSP70, and HSP90 interact with each other in HepG2.2.15 cells, suggesting that these HSPs function together in complex pathways. Because HSP60 interacts with human HBV pol and duck hepatitis B virus pol function relies on HSP90 and HSP70, we propose that HSP90 and HSP70 may be involved in the HBV life cycle with HSP60. Results of RNA interference experiments reported here are consistent with this hypothesis. Furthermore down-regulation of HSP90 or HSP70 results in the reduction of the HBV genome equivalents HBsAg and HBeAg secreted by HepG2.2.15 cells but does not affect cell proliferation or apoptosis. HSPs may interact with HBV pol protein to facilitate the formation of ribonucleoprotein complex and then the secretion of virus particles. Reversely the down-regulation of the HSPs may result in the decrease of viral RNPs and then affect the production of HBV. These studies are the first to demonstrate that HSP90, HSP70, and HSP60 can form a protein complex that plays a key role in regulating HBV replication and assembly.
Because these studies demonstrate that HSP90 plays a key role in the HBV life cycle, these results also suggest that HSP90 may be a putative therapeutic target for the treatment of HBV-related diseases. HSP90, with the development of 17-AAG, has emerged as a potential therapeutic target for cancer treatment (42). In this report, we demonstrate that treatment of HepG2.2.15 cells with 17-AAG resulted in the inhibition of HBV replication, suggesting that HSP90 may be a novel target for antiviral therapy.
In conclusion, this study involved a comprehensive, comparative proteomics analysis of the difference in cellular protein complexes between HepG2 and HepG2.2.15 cells using 2D BN/SDS-PAGE that resulted in the identification of a number of HBV-interacting proteins in two novel complexes. Furthermore we conclusively demonstrated, for the first time, that HSP90 and HSP70, with HSP60, as chaperone machinery are key modulators of the human HBV life cycle. Collectively these results suggest that BN/SDS-PAGE can be used to analyze functional protein complexes and that HSP90 and HSP70 are candidates for antiviral therapy.
Footnotes
Published, MCP Papers in Press, November 4, 2008, DOI 10.1074/mcp.M800250-MCP200
The abbreviations used are: HBV, hepatitis B virus; 17-AAG, 17-allylamino-17-demethoxygeldanamycin; 2D, two-dimensional; BN, blue native; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; MTT, methyl thiazolyl tetrazolium; RNP, ribonucleoprotein; siRNA, small interfering RNA; HSP, heat shock protein; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane- 1,3-diol; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; nt, nucleotide(s); pol, polymerase; ABP, actin-binding protein; BiP, immunoglobulin heavy chain-binding protein.
This work was supported in part by the Chinese State High-tech Program (863) (Grant 2006AA02A308), Chinese State Key Projects for Basic Research (973) (Grants 2006CB910401, 2006CB910801, and 2006CB910600), National Natural Science Foundation of China (Grants 30700988 and 30700356), National Natural Science Foundation of China for Creative Research Groups (Grant 30621063), and Beijing Municipal Key Project (Grant H030230280410).
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Ganem, D., and Prince, A. M. ( 2004) Hepatitis B virus infection—natural history and clinical consequences. N. Engl. J. Med. 350, 1118–1129 [DOI] [PubMed] [Google Scholar]
- 2.Beasley, R. P., Lin, C.-C., Hwang, L. Y., and Chen, C.-S. ( 1981) Hepatocellular carcinoma and hepatitis B virus: a prospective study of 22,707 men in Taiwan. Lancet 2, 1129–1133 [DOI] [PubMed] [Google Scholar]
- 3.Seeger, C., and Mason, W. S. ( 2000) Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64, 51–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paran, N., Geiger, B., and Shaul, Y. ( 2001) HBV infection of cell culture: evidence for multivalent and cooperative attachment. EMBO J. 20, 4443–4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sells, M. A., Chen, M. L., and Acs, G. ( 1987) Production of hepatitis B virus particles in HepG2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. U. S. A. 84, 1005–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Acs, G., Sells, M. A., Purcell, R. H., Price, P., Engle, R., Shapiro, M., and Popper, H. ( 1987) Hepatitis B virus produced by transfected Hep G2 cells causes hepatitis in chimpanzees. Proc. Natl. Acad. Sci. U. S. A. 84, 4641–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alberts, B. ( 1998) The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 92, 291–294 [DOI] [PubMed] [Google Scholar]
- 8.Ahlquist, P., Noueiry, A. O., Lee, W. M., Kushner, D. B., and Dye, B. T. ( 2003) Host factors in positive-strand RNA virus genome replication. J. Virol. 77, 8181–8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schägger, H., and Von Jagow, G. ( 1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 10.Wittig, I., Braun, H.-P., and Schägger, H. ( 2006) Blue native PAGE. Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 11.Camacho-Carvajal, M. M., Wollscheid, B., Aebersold, R., Steimle, V., and Schamel, W. W. ( 2004) Two-dimensional blue native/SDS gel electrophoresis of multiprotein complexes from whole cellular lysates. Mol. Cell. Proteomics 3, 176–182 [DOI] [PubMed] [Google Scholar]
- 12.Brouillard, F., Bensalem, N., Hinzpeter, A., Tondelier, D., Trudel, S., Gruber, A. D., Ollero, M., and Edelman, A. ( 2005) Blue native/SDS PAGE analysis reveals reduced expression of the mClCA3 protein in cystic fibrosis knock-out mice. Mol. Cell. Proteomics 4, 1762–1775 [DOI] [PubMed] [Google Scholar]
- 13.Farhoud, M. H., Wessels, H. J., Steenbakkers, P. J., Mattijssen, S., Wevers, R. A., van Engelen, B. G., Jetten, M. S., Smeitink, J. A., van den Heuvel, L. P., and Keltjens, J. T. ( 2005) Protein complexes in the archaeon Methanothermobacter thermautotrophicus analyzed by blue native/SDS-PAGE and mass spectrometry. Mol. Cell. Proteomics 4, 1653–1663 [DOI] [PubMed] [Google Scholar]
- 14.Stenberg, F., Chovanec, P., Maslen, S. L., Robinson, C. V., Ilaq, L. L., von Heijne, G., and Daley, D. O. ( 2005) Protein complexes of the Escherichia coli cell envelope. J. Biol. Chem. 280, 34409–34419 [DOI] [PubMed] [Google Scholar]
- 15.Ying, W., Jiang, Y., Guo, L., Hao, Y., Zhang, Y., Wu, S., Zhong, F., Wang, J., Shi, R., Li, D., Wan, P., Li, X., Wei, H., Li, J., Wang, Z., Xue, X., Cai, Y., Zhu, Y., and He, F. ( 2006) A dataset of human fetal liver proteome identified by subcellular fractionation and multiple protein separation and identification technology. Mol. Cell. Proteomics 5, 1703–1707 [DOI] [PubMed] [Google Scholar]
- 16.Lupberger, J., Mund, A., Kock, J., and Hildt, E. ( 2006) Cultivation of HepG2.2.15 on Cytodex-3: higher yield of hepatitis B virus and less subviral particles compared to conventional culture methods. J. Hepatol. 45, 547–552 [DOI] [PubMed] [Google Scholar]
- 17.Park, S. G., and Jung, G. ( 2001) Human hepatitis B virus polymerase interacts with the molecular chaperonin Hsp60. J. Virol. 75, 6962–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young, R. A. ( 1990) Stress proteins and immunology. Annu. Rev. Immunol. 8, 401–420 [DOI] [PubMed] [Google Scholar]
- 19.Jindal, S., and Young, R. A. ( 1992) Vaccinia virus infection induces a stress response that leads to association of Hsp70 with viral proteins. J. Virol. 66, 5357–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagawa, S., Umehara, T., Matsuda, C., Kuge, S., Sudoh, M., and Kohara, M. ( 2007) Hsp90 inhibitors suppress HCV replication in replicon cells and humanized liver mice. Biochem. Biophys. Res. Commun. 353, 882–888 [DOI] [PubMed] [Google Scholar]
- 21.Hu, J., and Seeger, C. ( 1996) Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl. Acad. Sci. U. S. A. 93, 1060–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Craig, E. A., Gambill, B. D., and Nelson, R. J. ( 1993) Heat shock proteins: molecular chaperones of protein biogenesis. Microbiology 57, 402–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itoh, H., Kobayashi, R., Wakui, H., and Komatsuda, A. ( 1995) Mammalian 60-kDa stress protein (chaperonin homolog). Identification, biochemical properties, and localization. J. Biol. Chem. 270, 13429–13435 [DOI] [PubMed] [Google Scholar]
- 24.Soltys, B. J., and Gupta, R. S. ( 1996) Immunoelectron microscopic localization of the 60-kDa heat shock chaperonin protein (Hsp60) in mammalian cells. Exp. Cell Res. 222, 16–27 [DOI] [PubMed] [Google Scholar]
- 25.Soltys, B. J., and Gupta, R. S. ( 1999) Mitochondrial-matrix proteins at unexpected locations: are they exported? Trends Biochem. Sci. 24, 174–177 [DOI] [PubMed] [Google Scholar]
- 26.Park, S. G., Lee, S. M., and Jung, G. ( 2003) Antisense oligodeoxynucleotides targeted against molecular chaperonin Hsp60 block human hepatitis B virus replication. J. Biol. Chem. 278, 39851–39857 [DOI] [PubMed] [Google Scholar]
- 27.Hu, J., Toft, D. O., and Seeger, C. ( 1997) Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 16, 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu, J., Toft, D., Anselmo, D., and Wang, X. ( 2002) In vitro reconstitution of functional hepadnavirus reverse transcriptase with cellular chaperone proteins. J. Virol. 76, 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duclos-Vallee, J. C., Capel, F., Mabit, H., and Petit, M. A. ( 1998) Phosphorylation of the hepatitis B virus core protein by glyceraldehydes-3-phosphate dehydrogenase protein kinase activity. J. Gen. Virol. 79, 1665–1670 [DOI] [PubMed] [Google Scholar]
- 30.Kann, M., and Gerlich, W. H. ( 1994) Effect of core phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J. Virol. 68, 7993–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouwmeester, T., Bauch, A., Ruffner, H., Angrand, P. O., Bergamini, G., Croughton, K., Cruciat, C., Eberhard, D., Gagneur, J., Ghidelli, S., Hopf, C., Huhse, B., Mangano, R., Michon, A. M., Schirle, M., Schlegl, J., Schwab, M., Stein, M. A., Bauer, A., Casari, G., Drewes, G., Gavin, A. C., Jackson, D. B., Joberty, G., Neubauer G., Rick, J., Kuster, B., and Superti-Furga, G. ( 2004) A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nat. Cell Biol. 6, 97–105 [DOI] [PubMed] [Google Scholar]
- 32.Mili, S., and Pinol-Roma, S. ( 2003) LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol. Cell. Biol. 23, 4972–4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feldmann, E., Schmiemann, V., Goedecke, W., Reichenberger, S., and Pfeiffer, P. ( 2000) DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 28, 2585–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertinato, J., Tomlinson, J. J., Schild-Poulter, C., and Haché, R. J. ( 2003) Evidence implicating Ku antigen as a structural factor in RNA polymerase II-mediated transcription. Gene ( Amst.) 302, 53–64 [DOI] [PubMed] [Google Scholar]
- 35.Li, L., Olvera, J. M., Yoder, K. E., Mitchell, R. S., Butler, S. L., Lieber, M., Martin, S. L., and Bushman, F. D. ( 2001) Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20, 3272–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang, C. J., Chen, Y. H., and Ting, L. P. ( 2000) Hepatitis B virus core protein interacts with the C-terminal region of actin-binding protein. J. Biomed. Sci. 7, 160–168 [DOI] [PubMed] [Google Scholar]
- 37.Henry, T., Gorvel, J.-P., and Meresse, S. ( 2006) Molecular motors hijacking by intracellular pathogens. Cell. Microbiol. 8, 23–32 [DOI] [PubMed] [Google Scholar]
- 38.Hill, V. M., Harmon, S. A., and Summers, D. F. ( 1986) Stimulation of vesicular stomatitis virus in vitro RNA synthesis by microtubule-associated proteins. Proc. Natl. Acad. Sci. U. S. A. 83, 5410–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buchkovich, N. J., Maquire, T. G., Yu, Y., Paton, A. W., Paton, J. C., and Alwine, J. C. ( 2008) Human cytomegalovirus specifically controls the levels of the endoplasmic reticulum chaperone BiP/GRP78, which is required for virion assembly. J. Virol. 82, 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung, J., Yueh, A., Appah, F. S., Jr., Yuan, B., de los Santos, K., and Goff, S. P. ( 2006) Interaction of Moloney murine leukemia virus matrix protein with IQGAP. EMBO J. 25, 2155–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Owsianka, A. M., and Patel, A. H. ( 1999) Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 257, 330–340 [DOI] [PubMed] [Google Scholar]
- 42.Sydor, J. R., Normant, E., Pien, C. S., Porter, J. R., Ge, J., Grenier, L., Pak, R. H., Ali, J. A., Dembski, M. S., Hudak, J., Patterson, J., Penders, C., Pink, M., Read, M. A., Sang, J., Woodward, C., Zhang, Y., Grayzel, D. S., Wright, J., Barrett, J. A., Palombella, V. J., Adams, J., and Tong, J. K. ( 2006) Development of 17-allylamino-17-demethoxygeldanamycin hydroquinone hydrochloride (IPI-504), an anti-cancer agent directed against Hsp90. Proc. Natl. Acad. Sci. U. S. A. 103, 17408–17413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, L., Xing, G., Tie, Y., Tang, Y., Tian, C., Li, L., Sun, L., Wei, H., Zhu, Y., and He, F. ( 2005) Role for the pleckstrin homology domain-containing protein CKIP-1 in AP-1 regulation and apoptosis. EMBO J. 24, 766–778 [DOI] [PMC free article] [PubMed] [Google Scholar]