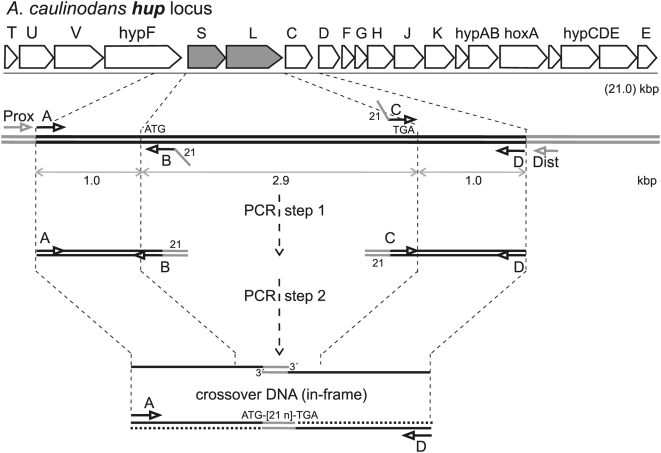
Figure 1. A. caulinodans hup genetic locus and physical map: creation of in-frame translational fusion deletions.
The top line represents the genetic map of the 20-gene hup polycistronic operon spanning 21 kbp. The second line represents an expanded physical map of hupSL DNA indicating postions of and (5′→3′) polarity for synthetic A, B, C, and D oligodeoxynucleotide primers of genome-identical sequence used in two, separate PCR reactions to generate DNA fragments A→B and C→D. The third line indicates a follow-up PCR reaction in which DNA fragments A→B and C→D were mixed, thermally denatured, allowed to partially renature, and used as combination PCR template/primer. As synthetic primers B and C share a complementary 21 bp extension sequence (angled line), the A→B Watson and C→D Crick strands (and vice versa) may partially reanneal via this 21 bp linker sequence In the third line, when such occurs, the resulting, partially-reannealed A→B(21 bp)C→D spliced DNA fragment which carries 5′-overhangs and free 3′-ends on both strands is a template for the thermostable DNA polymerase elongation reaction, producing a finished A→D duplex DNA fragment which may then be further amplified by PCR in the presence of added A and D primers. As verified by DNA sequencing analysis, finished, amplified A→D duplex fragments carry a genetic crossover which fuses (via the 21 bp linker sequence) in-frame the “start” codon of the proximal hupS gene to the “stop” codon of the distal hupL gene. Primers A and D may be extended with genome non-complementary elements to facilitate molecular cloning of resulting A→D fragments (Table 1). In vivo using homologous genetic recombination, wild-type loci are then exchanged for recombinant A→D crossover DNA fragments, which yield in-frame, translational deletion alleles of target genes of interest (Methods).