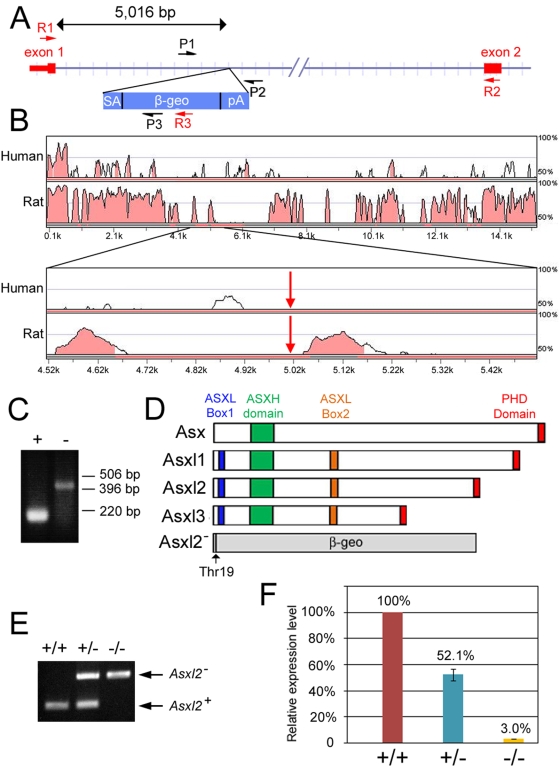
Figure 1. Generation of Asxl2 mutant mice.
(A) Schematic representation of the gene trap allele of murine Asxl2 (not drawn to scale). Exon 1 contains the 5'UTR (the narrow portion of the red box) and coding sequence for the first 19 amino acids of Asxl2. The gene trap cassette (blue rectangle) is inserted 5,016 bp downstream of exon 1. P1, P2 and P3: primers used in genotyping PCRs. R1, R2 and R3: primers used in RT-PCR analysis of the wild-type and gene trapped transcripts. (B) Conservation between mouse Asxl2 intron 1 and corresponding sequences in human ASXL2 and rat Asxl2 loci. The mouse, human and rat sequences were compared by SLAGAN. The 15,261-bp intron 1 of mouse Asxl2 was used as the base sequence. The top two panels show distribution of conserved sequence modules within the entire length of mouse Asxl2 intron 1. The bottom two panels show close-up views of 500 bp upstream and 500 bp downstream of the gene trap insertion site, which is indicated by red arrows. (C) RT-PCR analysis of the AQ0356 gene trap ES cell line. Transcripts from the wild-type (+) and mutant (−) alleles are detected with primer sets R1/R2 and R1/R3, respectively. The RT-PCR product in the mutant lane reflects splicing event that joins exon 1 with β-geo. (D) Schematic representation of the domain structures of Drosophila Asx, vertebrate Asxl1, Asxl2, Asxl3 and the predicted protein product from the Asxl2− allele. The ASXH (green box) and PHD (red box) domains are conserved from flies to mammals. The ASXL1 boxes 1 and 2 (blue and orange boxes) are conserved in the three mammalian Asx-like proteins but are not present in Asx. The mutant protein contains the first 19 amino acids of Asxl2 joined to β-geo. None of the conserved domains is present in the mutant protein. (E) PCR genotyping of genomic DNA from Asxl2 wild-type (+/+), heterozygous (+/−) and mutant (−/−) mice. P1 and P2 generate a 250-bp product from the wild-type allele. P1 and P3 generate a 480-bp product from the mutant allele. (F) Real-time RT-PCR quantification of wild-type Asxl2 transcripts in Asxl2 wild-type (+/+), heterozygous (+/−) and mutant (−/−) hearts. The transcript levels in heterozygous and mutant hearts were 52.1% and 3.0% of that in wild-type hearts, respectively.