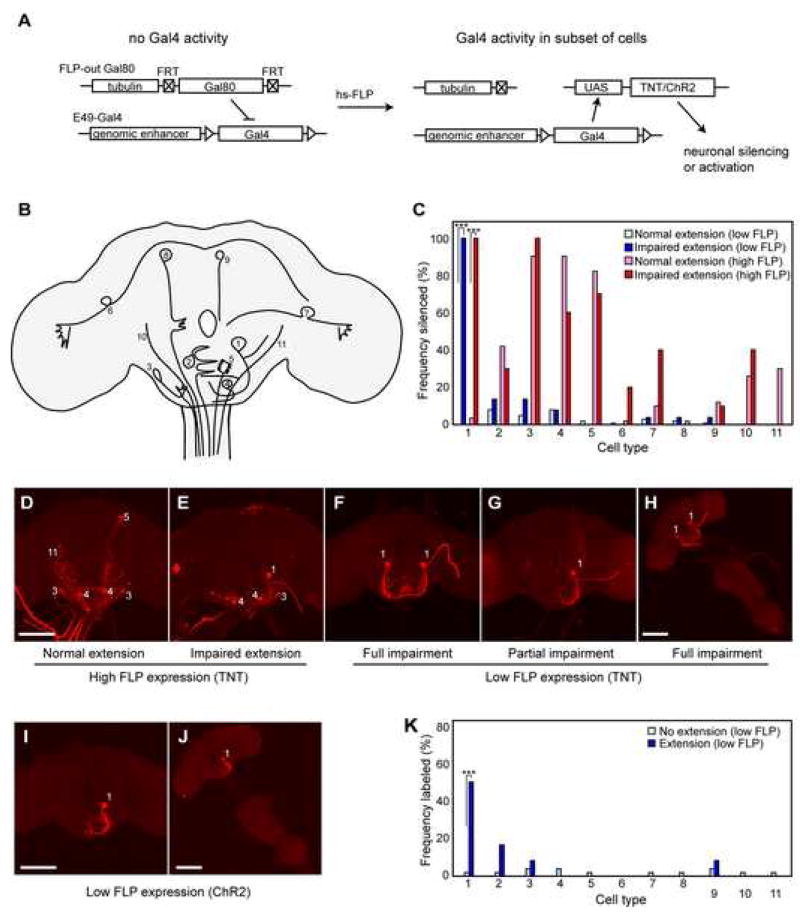
Figure 4. Mosaic analysis reveals neurons in E49-Gal4 population responsible for behavior.
(A) Mosaic silencing strategy using FLP-out Gal80. Prior to FLP expression, E49-Gal4 activity is repressed by ubiquitous expression of Gal80 repressor. Heat shock-driven FLP expression induces excision of the Gal80 cassette in a subset of cells, thereby allowing expression of tetanus toxin (UAS-TNT) and a visible marker (UAS-dsRed) (not illustrated) in those cells.
(B) Illustration of 11 cell types observed in FLP-out labeling during behavioral experiments.
(C) Frequencies of labeled (and therefore silenced) cell types observed in flies showing normal extension after low FLP expression (light blue bars; n=100 sampled from 1549 animals), impaired extension following low FLP expression (dark blue bars; n=68), normal extension following high FLP expression (pink bars; n=50 sampled from 220 animals), or impaired extension following high FLP expression (red bars; n=10), (genotypes: high FLP: hs-FLP; E49-Gal4/UAS-TNT; tub>Gal80>/UAS-dsRed; low FLP: tub>Gal80>; E49-Gal4/UAS-TNT; MKRS, hsFLP/UAS-dsRed). Frequencies represent the proportion of flies in each class where one or more cells of the given type have been silenced. Fisher’s exact test: *** = p<10−12. Cutoff for significance was set at 0.005 to produce an expected false positive rate of 5% of data sets (each including 11 cell types).
(D,E) Immunofluorescent detection of dsRed in example brains exposed to high FLP expression. The brain shown in (D) is from a fly showing normal extension; the brain in (E) is from a fly showing impaired extension. Numbers identify neuron types according to scheme shown in (B).
(F–H) Brains (F,G) or the complete CNS (H) of flies showing impaired PER following low FLP expression. Scale bars are 100μm.
(I,J) Representative images of brain (I) or full CNS (J) from flies showing rostrum extension in response to blue light following expression of dsRed (and hence ChR2) in neuron #1. Genotype: tub>Gal80>; E49-Gal4/UAS-ChR2; MKRS, hsFLP/UAS-dsRed.
(K) Frequency distributions of labeled (and therefore activated) cell types observed in flies showing no rostrum extension following exposure to blue light (light blue bars; n=50 sampled from 314 total) or extension of the rostrum (dark blue bars; n=12). Fisher’s exact test: *** = p<10−4. Cutoff for significance was set at 0.005 to produce an expected false positive rate of 5% of data sets (each including 11 cell types). From 314 animals screened, 12 showed extension of the rostrum. The false positives observed can be explained by the low level of background extension seen in the absence of Gal4 expression (figure 2).