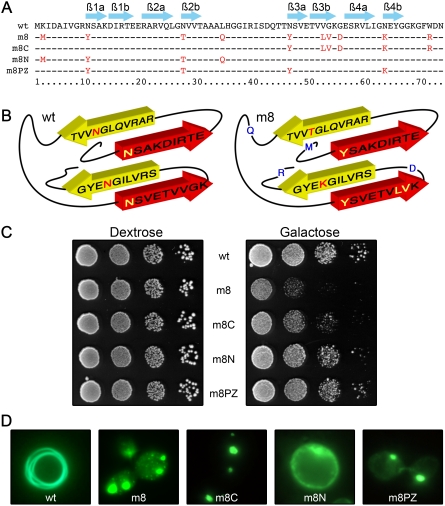
Figure 2. Identification of mutations required to induce toxicity.
(A) Amino acid sequence alignment of wild-type, m8 and chimeric mutants of HET-s(PrD) domains. Arrows above the sequence outline the ß-strands. (B) 3D modeling of HET-s(PrD) domain. Mutated amino acid in m8 (right panel) and wild-type (left panel) are located on a basic 3D model according to the RMN structure prediction [12]. (C) A ten-fold serial dilution of the various transformants was placed on solid dextrose or galactose medium and the growth of cells was observed after 48 h. (D) Mutants exhibit different GFP aggregation patterns in yeast. Cells were examined under a fluorescence microscope after 72 h of growth in liquid galactose medium.