Abstract
Chlamydophila pneumoniae, an obligate intracellular eubacterium, changes its form from a vegetative reticulate body into an infectious elementary body during the late stage of its infection cycle. Comprehension of the molecular events in the morphological change is important to understand the switching mechanism between acute and chronic infection, which is deemed to relate to the pathogenesis of atherosclerosis. Herein, we have attempted to screen genes expressed in the late stage with a genome-wide DNA microarray, resulting in nomination of 17 genes as the late-stage genes. Fourteen of the 17 genes and six other genes predicted as late-stage genes were confirmed to be up-regulated in the late stage with a quantitative reverse transcriptase–polymerase chain reaction. These 20 late-stage genes were classified into two groups by clustering analysis: ‘drastically induced’ and ‘moderately induced’ genes. Out of eight drastically induced genes, four contain σ28 promoter-like sequences and the other four contain an upstream common sequence. It suggests that besides σ28, there are certain up-regulatory mechanisms at the late stage, which may be involved in the chlamydial morphological change and thus pathogenesis.
Key words: Chlamydophila pneumoniae, DNA microarray, genome, chlamydia, gene expression
1. Introduction
Chlamydia and Chlamydophila species cause serious health problems in both humans and animals. These obligatory intracellular bacteria have apparently two distinct forms: an extracellular, infectious, and unproliferative elementary body (EB) and an intracellular, proliferative, and uninfectious reticulate body (RB). The beginning of infection is an attachment and internalization of EBs into host cells, followed by differentiation to RBs, defined as an early stage. RBs proliferate by binary fission in an inclusion at the mid-stage. After the proliferation, RBs change back to EBs at the late stage, and finally EBs are released from host cells for the next infection cycle. Accompanied with the morphological change during the late stage, chlamydial transcription and DNA replication are inactivated, and the nucleoid is condensed like heterochromatin of eukaryotic cells.
Chlamydophila pneumoniae is a causative agent of pneumonia and bronchitis of human1 and may contribute to the pathogenesis of atherosclerosis.2 To understand the pathogenesis, especially the mechanism of persistent infection, it is important to elucidate the molecular events in chlamydial development from RBs to EBs during the late stage, which may be related to the pathogenesis of atherosclerosis.3 The morphological conversion at the late stage is unique in chlamydia and thus the contributing genes could be targets for anti-chlamydosis therapy.
Chlamydiae have eukaryotic-type histone-like proteins, Hc1 and Hc2, encoded by hctA and hctB genes, respectively. Both proteins are produced specifically during the late stage and known to be associated with the concentrated nucleoid of EB.4–10 Promoter of hctB is recognized by σ28, one of the alternative sigma factors in Chlamydia.11 Thus, σ28 is thought to play a role in late expression of genes in the developmental cycle. Recently, it was reported that tlyC_1, bioY, dnaK, fmu, and pgk genes are recognized by chlamydial σ28 in vitro.12 The omcAB operon, ltuA, and ltuB genes are also known as late genes.13
So far there are two reports showing gene expression profile of Chlamydophila trachomatis during its development using a DNA microarray.14,15 Seventeen genes, including the known late genes such as hctB, ltuB, omcA, and omcB, are commonly annotated as late genes. However, the reports include discrepancies in expression annotation for many genes, especially for early-stage genes. It may indicate a technical difficulty to analyze chlamydial transcriptome in the early and mid phases of infection with DNA microarray. In spite of these intensive researches, molecular mechanism of chlamydial development is still unclear. In the present study, we developed C. pneumoniae DNA microarray covering nearly all genes on the genome of C. pneumoniae J13816 and focused on analysis of the mid to late stage of chlamydial gene expression.
2. Materials and methods
2.1. Cell and chlamydia culture
Infection, culture, and DNA preparation of C. pneumoniae J138 were performed as previously described.17 Briefly, HEp-2 cells (ATCC CCL-23), a human epithelioid larynx carcinoma-derived cell line, were maintained using Iscove's modified Dulbecco's medium (Gibco BRL, CA, USA) supplemented with 10% heat-inactivated fetal calf serum (FCS) (PAA Laboratories, Inc., USA) and 10 µg gentamicin per milliliter. HEp-2 cells grown in 6-well tissue culture plates for 24 h after passage were infected with 1 × 106 inclusion forming unit (IFU) of C. pneumoniae per well, resulting in multiplicity of infection of 0.5. The inoculum was centrifuged at 700g for 1 h at 22°C followed by incubation at 36°C with 5% CO2 for one more hour. After the extracellular bacteria were removed, infected cells were further incubated in the same medium without 5% of FCS and addition of 1.0 µg/mL cycloheximide (Sigma, St. Louis, MO, USA). Growth curve of C. pneumoniae was determined as IFU by counting inclusion bodies cultured on a 96-well plate as described previously.18 Genomic DNA was extracted from purified organisms with EZ1 DNA Tissue Kit (QIAGEN, Hilden, Germany).
2.2. C. pneumoniae DNA microarray
We developed a DNA microarray composed of 884 DNA fragments covering 986 genes, except 83, from the C. pneumoniae J138 genome. Of the 884 DNA fragments, 497 cover one chlamydial gene or 23S rDNA in each fragment, and the other 387 contain plural genes since a part of the DNA fragments were prepared using clones for random sequencing of C. pneumoniae J138 genome.16 Negative control DNA fragments are prepared with adk gene from Pseudomonas putida mt-2, and two human cDNA of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin. DNA fragments were prepared with polymerase chain reaction (PCR) and spotted to two positions on a microarray glass.
2.3. Preparation of fluorescent-labeled probes and DNA microarray analysis
Total RNA was purified from cells both infected and uninfected with C. pneumoniae J138 using RNeasy (QIAGEN). Chlamydial RNA was enriched by removing host mRNA and rRNA from the total RNA with magnetic beads of MicrobEnrich kit (Ambion, TX, USA).19 Fluorescent-labeled cDNA probe was prepared using RNA as the template with Atlas PowerScript Fluorescent Labeling Kit (BD Biosciences Clontech, CA, USA), Cy3, and Cy5 dye (Amersham). Fluorescent-labeled genomic DNA was prepared using 100 ng of purified chlamydial genomic DNA as the template with random prime method using BioPrime DNA Labeling System (Invitrogen, Carlsbad, CA, USA), dNTPs (Amersham, NJ, USA) and aminoaryl-dUTP (Sigma), instead of biotin-dCTP and Cy3 or Cy5 dye (Amersham). After PCR for aminoaryl-dUTP incoporation, the reaction was terminated with ethanol precipitation. DNA resuspended within 0.1 M sodium hydrogencarbonate was mixed with the same volume of Cy3 or Cy5 dye solution dissolved in DMSO. After incubation at 37°C for 1 h, labeled DNA probes were purified with QIAquick PCR purification kit (QIAGEN). This method provides fluorescent-labeled genomic DNA probe sufficient for two or more times of hybridization. When the genomic DNA probe was hybridized to the DNA microarray, 15 of total 1768 chlamydial spots were removed for further analysis due to under-detectable intensity.
Separately fluorescence-labeled cDNA probe and the genomic DNA were mixed and hybridized competitively to a microarray glass at 60°C for 16 h, as previously described.20 After washing, the glass was scanned with FLA-8000 (Fujifilm, Tokyo). Threshold of each experiment was determined as an average plus a value of standard deviation from negative spot intensities. Only spots with higher fluorescent intensity than the threshold were further analyzed. Gene expression value at each spot was calculated as a ratio of intensity of cDNA fluorescence against that of genomic DNA.
To normalize and compare data from each RNA preparation, ompA gene was adopted as a continuously expressing standard due to the following reasons. First, global normalization using neither mean nor median is not appropriate for our data, because the detection rate varies remarkably from the time of infection (Fig. 1A). Secondly, microarray spots for chlamydial 23S rRNA showed non-specific signals derived from host RNA, and the amount of chlamydial rRNA was affected by MicrobEnrich treatment which is unavoidable for a reliable chlamydial DNA microarray analysis (data not shown). Thirdly, gene expression pattern of the ompA was similar to one of the chlamydial ribosomal RNA based on the reverse transcriptase–PCR (RT–PCR) analysis when no MicrobEnrich treatment was carried out (Supplementary Fig. S2C). At last, the ompA gene was detected with the DNA microarray at all points analyzed, and the amount of the ompA gene transcript was not affected by treatment with MicrobEnrich (data not shown).
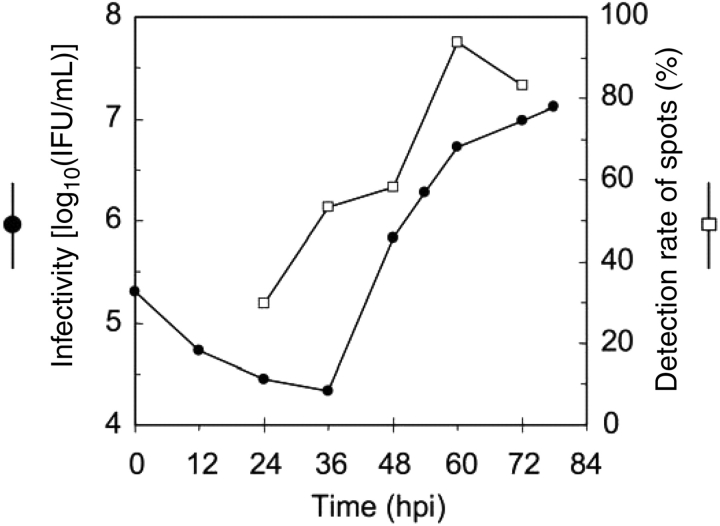
Figure 1.
Gene expression analysis with DNA microarray. Reproduction of infectious progenies of C. pneumoniae J138 (shown on left as ‘Infectivity’) and detection rate of spots (on right) by Cy3-labeled cDNA hybridization on DNA microarray. 1608 valid spots were chosen based on Cy5-labeled genome hybridization. Percentage of the spots with intensity over threshold, when the cDNA probe was hybridized, was represented. When dyes were flipped, similar results were obtained.
2.4. Quantitative RT–PCR
Quantitative RT–PCR was carried out with total RNA extracted from infected cells using QuantiTect SYBR Green RT–PCR Kit (QIAGEN) with LightCycler (Roche). In the RT–PCR, total RNA was directly submitted to RT reaction without the MicrobEnrich treatment. Therefore, signals of chlamydial rRNA were used as references, and with this rRNA reference, it was confirmed that the ompA gene is continuously expressed. RNA purified from the cells at 24, 36, 48, 60, and 72 h after infection was diluted appropriately for assay, and was mixed with primers and reagents. Primer sequences used in this work are summarized in Supplementary Table S1. The reaction was performed according to the manuals from suppliers. Expression amount of each gene was normalized with that of chlamydial 16S rRNA.
2.5. Clustering and statistical analysis
Data were normalized by median of each gene before clustering. Clustering analysis of the data was performed with ‘Cluster’,21 and the results were viewed with ‘TreeView’. Statistical comparisons were made using Wilcoxon's t-test and Mann–Whitney's U-test for analysis of the DNA microarray data and Chi square test for gene deviation analysis. P-value of <0.05 were considered significant.
3. Results
3.1. Chlamydial DNA microarray analysis
To distinguish the mid and late stages in the intracellular development of C. pneumoniae J138, the timing of progeny production was examined prior to analyzing gene expression during differentiation from EBs to RBs. The number of re-infectious progenies starts to increase between 36 and 48 hours post-infection (hpi) of C. pneumoniae J138 to HEp-2 cells (Fig. 1). Thus, in this work, the late stages in developmental cycle were defined after 48 hpi.
When RNAs are prepared from chlamydia infection samples, amount of host RNA contamination is generally critical for data sensitivity and reliability. To reduce host cell RNA from total RNA, MicrobEnrich Kit was adopted to enrich chlamydial RNA before labeling, and the J138 genomic DNA was used as a counter control of the RNA to be tested. As with the RNA at 24 hpi, approximately 30% of chlamydial spots (total of 1768 spots by duplication of 884 DNA fragments covering 986 genes) exhibited higher fluorescence intensity than a threshold. At 60 hpi, 1508 spots were detected and the ratio of positive spots increased up to 93.8%, whereas 1608 spots were detected with genomic DNA as reference spots for cDNA (Fig. 1). The low detection ratio with RNAs from 24 hpi may be due to low concentration of chlamydial RNA in the total RNA, which still contains certain amount of the host RNA even after enrichment of chlamydial RNA using MicrobEnrich Kit. At earlier time point than 24 hpi fluorescent signals on the array were critically weak and the data were much less reliable than that after 24 hpi. Herein, we have focused on gene expression between mid and late stages (24–72 hpi).
3.2. Genome-wide gene expression analyses with chlamydial DNA microarray
Using chlamydial RNA-enriched RNA samples at 24–72 hpi, stage-specific gene expression was analyzed with external and internal controls, which were C. pneumoniae J138 genomic DNA and ompA gene product, respectively. The spots without genomic DNA signals were eliminated from the data analyses as detection errors. Relatively strong and constitutive ompA gene expression was adopted as an internal control to normalize individual experiments of DNA microarray analysis (for detail, see Section 2). Out of 1768 spots duplicated by 884 DNA fragments carrying a total of 986 genes of C. pneumoniae J138, the amount of detected spots were varied at 24–72 hpi, such as only 30% of whole spots were detected at 24 hpi while the maximum detection rate, 85% (1508 positive spots), was observed using 60 hpi RNA sample (Fig. 1). In order to screen late-stage-specific genes, duplicated spots for each DNA fragment were treated as two individual spots, and a clustering analysis was performed using the data of approximately 400 out of 1768 spots, intensities of which were higher than threshold at any time point (Fig. 2A).
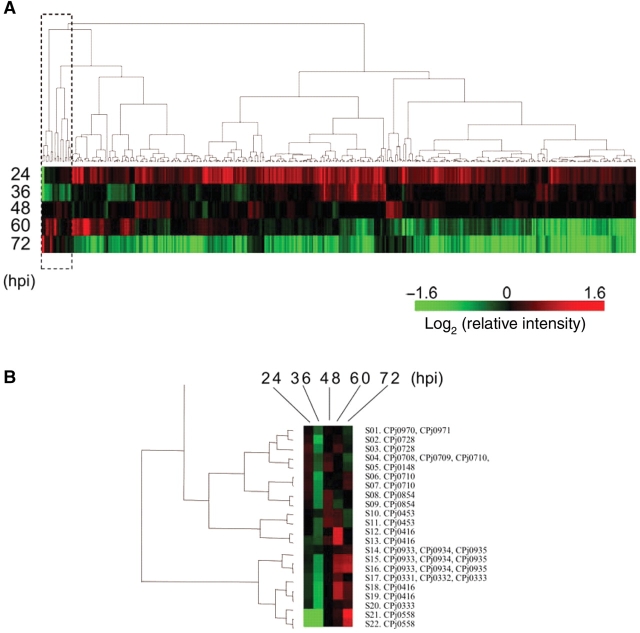
Figure 2.
Clustering analysis of microarray data. (A) The data were normalized with ompA expression as described under Section 2. A hierarchical cluster was obtained by analysis of 440 data. (B) A magnified view of a cluster in a rectangle with a broken line in the (A). Spot names (S01–S23) and gene IDs (CPjXXXX) are represented on the right side.
The result from the clustering analysis indicates that expression of most genes tends to diminish from mid to late stage of infection. Approximately a half of the genes is drastically down-regulated, and the other half of the genes is constitutive or gradually decreased. In contrast, 22 spots, including duplicated spots for same DNA fragments, exhibited an upward trend with respect to time as a cluster collected on one side of the clustering result (box in Fig. 2A and B). A total of 17 genes in the 22 spots were assigned as candidates of the late genes and compared with results reported previously, including a proteomic analysis in Table 1.
Table 1.
Summary of DNA microarray and RT–PCR analyses
| Gene ID | Name | Definition | Microarray | RT–PCR | Mukhopadhyay et al.a | CTb | Nicholson et al.a | Belland et al.a |
|---|---|---|---|---|---|---|---|---|
| CPj0331 | – | CT082 hypothetical protein | Late | I | ND | CT082 | Late | ND |
| CPj0332 | – | CHLTR T2 protein | Late | I | ND | – | ||
| CPj0333 | ltuB | LtuB protein | Late | I | ND | CT080 | Late | Late |
| CPj0558 | omcA | 9 kDa cysteine-rich lipoprotein | Late | I | ND | CT444 | Late | Late |
| CPj0416 | himD | Integration host factor alpha | Late | II | ND | CT267 | Midlate-I | ND |
| CPj0453 | pmp14 | Polymorphic outer membrane protein H family | Late | II | ND | CT872 | Midlate-I | Late |
| CPj0708 | – | CT668 hypothetical protein | Late | II | ND | CT668 | Midlate-I | ND |
| CPj0709 | – | CT667 hypothetical protein | Late | II | ND | CT667 | Midlate-I | ND |
| CPj0710 | – | CT666 hypothetical protein | Late | II | Up | CT666 | Midlate-I | ND |
| CPj0728 | – | CHLPN 76 kDa homolog_1 (CT622) | Late | II | ND | CT622 | Late | ND |
| CPj0854 | ompB | Outer membrane protein B | Late | II | ND | CT713 | Midlate-II | ND |
| CPj0933 | – | Disulfide bond isomerase | Late | II | ND | CT783 | Midlate-II | Late |
| CPj0970 | yccA2 | Transport permease | Late | II | ND | CT819 | Midlate-I | ND |
| CPj0971 | ftsY | Cell division protein ftsY | Late | II | ND | CT820 | Constitutive | ND |
| CPj0148 | – | S/T protein kinase | Late | Const. | ND | CT145 | Constitutive | ND |
| CPj0934 | rnpA | Ribonuclease P protein component | Late | Const. | ND | CT784 | Constitutive | ND |
| CPj0935 | rl34 | L34 ribosomal protein | Late | Const. | ND | CT785 | Constitutive | ND |
| CPj0577 | – | SWIB (YM74) complex protein | NL | Const. | ND | CT460 | Constitutive | ND |
| CPj0695 | ompA | Major outer membrane protein | NL | Const. | Up | CT681 | Midlate-II | ND |
| CPj0769 | topA | DNA topoisomerase I-fused to SWI domain | NL | Const. | ND | CT643 | Late | ND |
| CPj0878 | set | SET domain protein | NL | Latec | ND | CT737 | Constitutive | ND |
| CPj0244 | adk | Adenylate kinase | NC | I | Up | CT128 | Constitutive | ND |
| CPj0384 | hctB | Histone-like protein 2 | NC | I | ND | CT046 | Late | Late |
| CPj0678 | – | Hypothetical protein | NC | I | ND | – | ||
| CPj0886 | hctA | Histone-like developmental protein | NC | I | ND | CT743 | Constitutive | Late |
| CPj0466 | pmp15 | Polymorphic outer membrane protein E family | NC | II | ND | CT869 | Late | ND |
| CPj0559 | – | CT444.1 hypothetical protein | NC | II | ND | CT444.1 | ND | ND |
| CPj0238 | zwf | Glucose-6-P dehyrogenase | NC | Const. | ND | CT185 | Constitutive | ND |
| CPj0679 | pgk | Phosphoglycerate kinase | NC | Const. | ND | CT693 | Late | ND |
Late, late-stage gene; NL, not-late-stage genes (mid or constitutive genes); NC, not clustered in Fig. 2A; I, class I; II, class II; Const., constitutive in Fig. 3; ND, not determined.
aThe classifications follow the cited reference.
bOrtholog of C. trashomatis represented as gene number. ‘ − ’, no ortholog.
cMurata et al (2006).
3.3. Identification of late genes by quantitative RT–PCR
Expression profiles of the 17 genes classified as late genes based on the DNA microarray were analyzed with quantitative RT–PCR using RNA harvested at 24, 36, 48, 60, and 72 hpi (Supplementary Fig. S2). Fourteen (82%) of the 17 genes were confirmed to be induced at the late stage (Table 1). However, the other three genes, CPj0148, CPj0934, and CPj0935, were constitutive in the RT–PCR analysis. The DNA fragment containing CPj0934 and CPj0935 for the DNA microarray also carries CPj0933, which is consistently categorized into the late gene (S14–S17 in Fig. 2B).
Expressions of other 11 genes, which were not categorized into the late genes in the microarray analysis, were also examined with quantitative RT–PCR. CPj0466 (pmp15, polymorphic outer membrane protein E family) and CPj0679 (pgk, phosphoglycerate kinase) were reported as late-stage genes in C. trachomatis gene expression and C. pneumoniae proteome analysis.14,22 CPj0244 (adk gene) was tested because the gene product adenylate kinase (AK) was reported to increase in protein amount in the RB–EB transition stage of C. pneumoniae.22 Four genes, CPj0878 (set, SET domain protein), CPj0769 (topA, DNA topoisomerase I-fused to SWI domain), CPj0886 (hctA, histone-like developmental protein), and CPj0384 (hctB, histone-like protein 2), which are partially similar to eukaryotic chromatin structure regulation or polycomb complex proteins, were also reported as late-stage genes10,15,22 Here, we tested one more gene, CPj0577 (SWIB complex protein), which is eukaryotic chromatin structure regulation or polycomb complex proteins. CPj0678 (hypothetical protein) and CPj0559 (CT444.1 hypothetical protein) were tested to distinguish their expression from adjacent genes CPj0679 and CPj0558, respectively. CPj0695 (ompA, major outer membrane protein) and CPj0238 (zwf, glucose-6-P dehyrogenase) were tested as controls for constitutive expression.
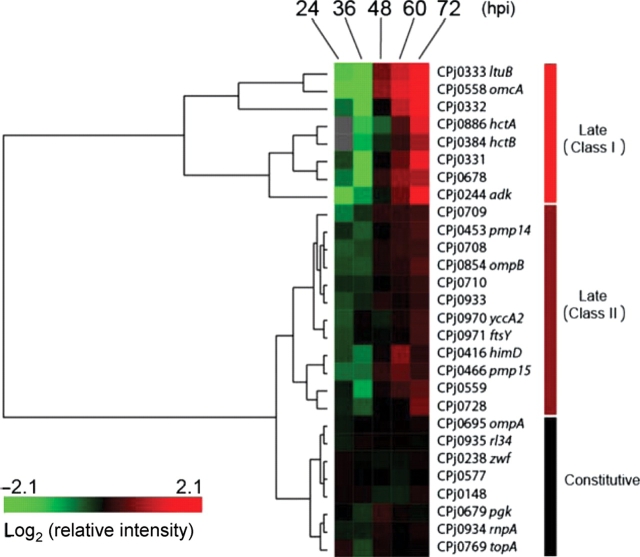
A total of 28 genes were examined with quantitative RT–PCR using RNA harvested at 24–72 hpi (Supplementary Fig. S2). These 28 genes were divided into three clusters based on the results of the clustering analysis (Fig. 3, Table 1, Supplementary Fig. S2A). Eight genes were categorized as class I genes that were induced sharply (more than five times induction ratio) at the late stage, 12 genes were class II genes induced moderately (from two to five times) at the late stage, and the other eight were constitutive genes. Out of the 20 late genes, hctA, hctB, adk, and CPj0710 are known as late genes in C. pneumoniae,10,22 and other 16 genes are identified as novel late genes.
Figure 3.
Clustering analysis of RT–PCR data. The relative expression data normalized with chlamydial 16S rRNA were subjected to clustering analysis to classify genes. Transcript of hctA and hctB genes was under detection limit at 24 hpi with RT–PCR. According to the dendrogram, genes were classified in three groups.
3.4. Conserved sequences in the late genes
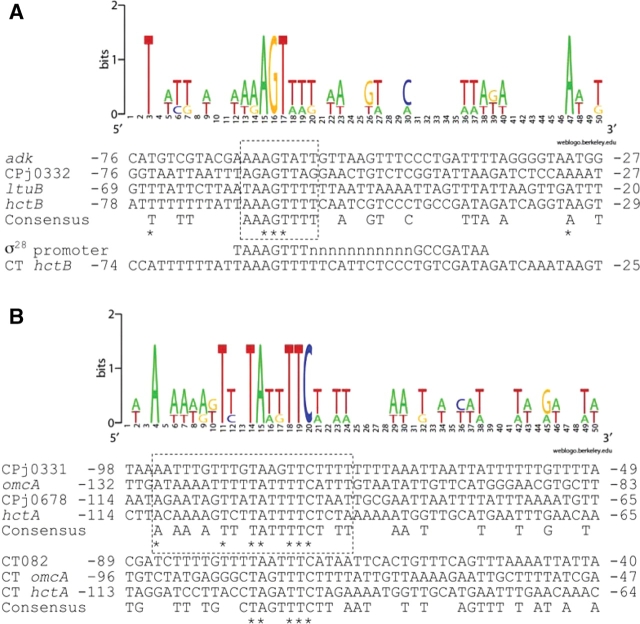
Chlamydial gene expression at the late stage has been analyzed using C. trachomatis, and it has been proposed that σ28 RNA polymerase is involved in the late gene expression.12,23,24 We compared upstream sequences of the C. pneumoniae late genes identified here. Four out of the eight class I genes, hctB, ltuB, CPj0332, and adk, contain similar sequences to the −35 consensus of Escherichia coli σ28 promoter at approximately 60 nucleotides upstream of each putative translation initiation site of gene25 (Fig. 4A). It has been reported that AT-rich sequence upstream of promoter is one of the σ28 preferences by Shen et al.24 In fact, these genes have an AT-rich sequence upstream of −35 consensus. These findings suggest that σ28 RNA polymerase transcribes these genes. However, the CGA motif for −10 σ28 promoter was not found except in the hctB gene, suggesting that the consensus sequence of chlamydial σ28 promoter is different from E. coli.
Figure 4.
Consensus sequence upstream of class I late genes. (A) Multiple alignment of upstream sequences of hctB, ltuB, CPj0332, and adk genes. They have a consensus sequence partially similar to σ28 promoter consensus sequence. (B). Multiple alignment of upstream sequences of CPj0331, CPj0678, omcA, and hctA genes of C. pneumoniae and the ortholog genes in C. trachomatis (CT). CT082 is the ortholgog of CPj0331. Number represents nucleotides from the putative translation initiation site. Consensus sequences are determined as three or more of four nucleotides are identical in C. pneumoniae genes, or two or more are identical in C. trachomatis. Asterisks represent conserved nucleotides in all genes in each species. Highly conserved regions in C. pneumoniae genes are boxed with dashed lines. Sequence logos are generated with WebLogo (http://weblogo.berkeley.edu/) using aligned sequences of C. pneumoniae genes.
The other four genes in class I, CPj0331, CPj0678, omcA, and hctA, contain no sequences similar to the typical σ28 promoter, but a conserved AT-rich sequence, AnAAnAnTTnTATTTTCTnTT, approximately 100 nucleotides upstream from each predicted gene start site (Fig. 4B). The conserved AT-rich sequence was not found in other class I, class II, and constitutive genes determined with RT–PCR. Moreover, a part of the AT-rich sequence, such as TAnnTTC, was also found upstream of the orthologous genes of C. trachomatis. It may suggest that the AT-rich sequence (or a part of it) is conserved in the distinctive chlamydial genera and may be involved in late gene expression of the orthologues.
Unlike hctB gene, adk and ltuB genes of C. trachomatis have no σ28 consensus sequence, even though ltuB gene of C. trachomatis is also a late gene. There is no orthologous gene of CPj0332 in C. trachomatis genomes. These differences between C. pneumoniae and C. trachomatis suggest two possibilities: one is that σ28 of these bacteria is capable of recognizing the distinct sequence and/or regulates the distinct set of genes, and the other is that there is a distinct regulation mechanism for late expression in each species.
The late genes, CPj0331, CPj0332, and ltuB (CPj0333), are tandem located on a same strand, separated by 220, 141, and 95 bp intergenic spaces upstream of these genes, respectively. Our results revealed that upstream non-coding regions of ltuB and CPj0332 contain σ28 promoter-like sequences, suggesting that these two promoters are individually recognized by σ28 RNA polymerase.
4. Discussion
Here we describe a transcriptome profile of C. pneumoniae in the late stage of the infection using DNA microarray and quantitative RT–PCR. Based on the DNA microarray screening, 17 C. pneumoniae genes were nominated as up-regulated genes in the late stage of infection, whereas approximately half of the genes analyzed are constitutively expressed through infection and the other half were down-regulated in the late stage. The 17 genes shown as late genes by the DNA microarray and 11 genes predicted as late genes based on its sequence and functional similarities to genes expressed in late stage in other chlamydial species were analyzed with quantitative RT–PCR. As a result, a total of 20 genes (14 out of the 17 genes and six of the 11 genes) were confirmed as late genes. Previously, we demonstrated that hctA, hctB, and set genes of C. pneumoniae are expressed at the late stage of infection.10 AK encoded by adk gene and product of Cpn0710, an orthologue of CPj0710, have been identified to be increased in the RB–EB transition stage of C. pneumoniae.22 Thus, except for these genes, we identified 16 novel late genes in this study. Expression of ompB, adk, and Cpn0710 genes are increased by interferon-γ (IFN-γ) treatment.26,27 These findings indicate that there are some mechanisms shared by molecular systems leading to the persistent infection (RBs to atypical RBs) induced by IFN-γ and to the late stage (RBs to EBs). The 20 late genes were divided into two classes by the clustering analysis: eight sharply induced genes (class I) and 12 moderately up-regulated genes (class II). The class I genes, hctA, hctB, omcA, ltuB, adk, CPj0678, CPj0331, and CPj0332, begin to be induced synchronously when the conversion from RBs to EBs starts at around 48 hpi. As hctA and hctB genes are believed to contribute to the transition of RBs to EBs, somehow other six class I genes may play their roles or explain the cellular status in the late stage.
Four genes (hctB, ltuB, adk, and CPj0332) out of eight class I genes contain the upstream sequences which may be recognized by σ28 RNA polymerase, supporting the hypothesis that σ28 RNA polymerase is responsible for late gene expression.12,23,24 However, the late gene, ltuB, of C. trachomatis does not have the σ28 consensus sequence, suggesting a distinct mechanism, which allows the late expression of the gene without σ28 in C. trachomatis.
In contrast, the other four genes of class I (CPj0331, omcA, CPj0678, and hctA) have no σ28 promoter-like sequence, suggesting the presence of other mechanism(s) of late expression. Instead, we found a common AT-rich sequence around −100 nucleotides upstream of them (Fig. 4B), suggesting that it plays a role in late expression. In addition, the sequence was also conserved partially upstream of the orthologous genes of C. trachomatis. This sequence is not similar to the known promoter sequences and far from each gene. In fact, the core TAnnTTC of C. trachomatis hctA gene is located at −45 to −51 from the transcription initiation site.13 Therefore, it might be a binding site of regulatory protein(s) rather than a promoter sequence.
AK catalyzes a reversible high-energy phosphoryl transfer reaction between adenosine triphosphate (ATP) and AMP to generate ADP and is considered to contribute to the homeostasis of cellular adenine nucleotide composition in the cell.17 There might be two possible reasons why the adk gene is up-regulated at the late stage of infection: one is that AK is necessary to increase ATP concentration or to decrease ADP concentration, and the other is that it is necessary to decrease AMP concentration. In the first case, AK produces ATP and AMP from ADP. Because a Km (Michaelis constant) value for AMP of the AK is remarkably higher than other AKs,17 accumulation of AMP seems not to affect the reaction. In the second case, ADP is produced to consume AMP through the opposite reaction. Because ADP is a substrate of chlamydial ADP/ATP translocase, it could promote to get ATP from the host cell. We cannot conclude here which is more likely. However, in many organisms intracellular adenine nucleotide balance is strictly regulated, and change of the balance induces various responses in the cell, including not only metabolism regulation, but also cell differentiation. In Bacillus subtilis increase of ADP (or low ATP/ADP ratio) is hypothesized to be a cell differentiation signal for sporulation.28 Therefore, also in C. pneumoniae the change of adenine nucleotide balance could induce not only adk gene, but also other late genes to differentiate RB into EB.
In the class II genes, himD gene encoding a subunit of histone-like protein integration host factor and pmp15 gene encoding outer membrane protein are up-regulated transiently at 60 hpi. This temporary enhancement of expression was consistent with the results of binding assay in vitro using an extract from C. trachomatis reported by Zhong et al.29 Expression of set gene encoding SET domain protein has been previously reported to be expressed during late stage accompanied with one of hctA and hctB genes,10 suggesting that it plays an important role in chlamydial development by the chlamydial histone-like protein modification.
Recently, OmcB protein of C. trachomatis, one of the products of late omcAB operon, has been revealed to be a surface-exposed adhesion to infect the host cell.30 Likewise, other membrane proteins encoded by late genes identified here, omcA, pmp14, pmp15, and ompB, might function to attach to the host cell.
In this study, we have identified 16 novel late genes of C. pneumoniae and predicted cis-acting sequences common in upstream regions of the late genes. The genome sequence of Chlamydia contained a small number of regulatory systems, a single pair of bacterial two-component system, and only several transcription factors. However, our results suggest that regulation of gene expression in C. pneumoniae might be more complicated than expected. Further investigation using a DNA microarray may help to understand chlamydial physiology and pathology.
Supplementary Data
Supplementary data are available online at www.dnaresearch.oxfordjournals.org.
Funding
This study was supported by the Japan Society for the Promotion of Science, Research for the Future Program (JSPS-RETF00L01411), and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (KAKEN-HI: 14770063 and 16790255).
Supplementary Material
Acknowledgements
We thank Taku Oshima (Nara Inst. Sci. Technol.) for his helpful suggestions and Ryutaro Fujinaga (Yamaguchi Univ.) who helped us to start this study.
References
- 1.Grayston J. T., Kuo C. C., Campbell L. A., et al. Chlamydia pneumoniae sp. Nov. for Chlamydia sp. Strain TWAR, Int. J. Syst. Bacteriol. 1989;39:88–90. [Google Scholar]
- 2.Kuo C. C., Shor A., Campbell L. A., et al. Demonstration of Chlamydia pneumoniae in atherosclerotic lesions of coronary arteries. J. Infect. Dis. 1993;167:841–849. doi: 10.1093/infdis/167.4.841. [DOI] [PubMed] [Google Scholar]
- 3.Ramirez J. A. Isolation of Chlamydia pneumoniae from the coronary artery of a patient with coronary atherosclerosis. The Chlamydia pneumoniae/Atherosclerosis Study Group. Ann. Intern. Med. 1996;125:979–982. doi: 10.7326/0003-4819-125-12-199612150-00008. [DOI] [PubMed] [Google Scholar]
- 4.Barry C. E., Hayes S. F., Hackstadt T. Nucleoid condensation in Escherichia coli that express a chlamydial histone homolog. Science. 1992;256:377–379. doi: 10.1126/science.256.5055.377. [DOI] [PubMed] [Google Scholar]
- 5.Hackstadt T., Baehr W., Ying Y. Chlamydia trachomatis developmentally regulated protein is homologous to eukaryotic histone H1, Proc. Natl. Acad. Sci. USA. 1991;88:3937–3941. doi: 10.1073/pnas.88.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perara E., Ganem D., Engel J. N. A developmentally regulated chlamydial gene with apparent homology to eukaryotic histone H1. Proc. Natl. Acad. Sci. USA. 1992;89:2125–2129. doi: 10.1073/pnas.89.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao S., Kaul R., Wenman W. M. Identification and nucleotide sequence of a developmentally regulated gene encoding a eukaryotic histone H1-like protein from Chlamydia trachomatis. J. Bacteriol. 1991;173:2818–2822. doi: 10.1128/jb.173.9.2818-2822.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brickman T. J., Barry C. E., 3rd, Hackstadt T. Molecular cloning and expression of hctB encoding a strain-variant chlamydial histone-like protein with DNA-binding activity. J. Bacteriol. 1993;175:4274–4281. doi: 10.1128/jb.175.14.4274-4281.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pedersen L. B., Birkelund S., Christiansen G. Interaction of the Chlamydia trachomatis histone H1-like protein (Hc1) with DNA and RNA causes repression of transcription and translation in vitro. Mol. Microbiol. 1994;11:1085–1098. doi: 10.1111/j.1365-2958.1994.tb00385.x. [DOI] [PubMed] [Google Scholar]
- 10.Murata M., Azuma Y., Miura K., et al. Chlamydial SET domain protein functions as a histone methyltransferase. Microbiology. 2007;153:585–592. doi: 10.1099/mic.0.29213-0. [DOI] [PubMed] [Google Scholar]
- 11.Yu H. H., Tan M. Sigma28 RNA polymerase regulates hctB, a late developmental gene in Chlamydia. Mol. Microbiol. 2003;50:577–584. doi: 10.1046/j.1365-2958.2003.03708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu H. H., Kibler D., Tan M. In silico prediction and functional validation of sigma28-regulated genes in Chlamydia and Escherichia coli. J. Bacteriol. 2006;188:8206–8212. doi: 10.1128/JB.01082-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fahr M. J., Douglas A. L., Xia W. Characterization of late gene promoters of Chlamydia trachomatis. J. Bacteriol. 1995;177:4252–4260. doi: 10.1128/jb.177.15.4252-4260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belland R. J., Nelson D. E., Virok D., et al. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc. Natl. Acad. Sci. USA. 2003;100:15971–15976. doi: 10.1073/pnas.2535394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholson T. L., Olinger L., Chong K., et al. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J. Bacteriol. 2003;185:3179–3189. doi: 10.1128/JB.185.10.3179-3189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shirai M., Hirakawa H., Kimoto M., et al. Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res. 2000;28:2311–2314. doi: 10.1093/nar/28.12.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miura K., Inouye S., Sakai K., et al. Cloning and characterization of adenylate kinase from Chlamydia pneumoniae. J. Biol. Chem. 2001;276:13490–13498. doi: 10.1074/jbc.M009461200. [DOI] [PubMed] [Google Scholar]
- 18.Yoneda H., Miura K., Matsushima H., et al. Aspirin inhibits Chlamydia pneumoniae-induced NF-kappa B activation, cyclo-oxygenase-2 expression and prostaglandin E2 synthesis and attenuates chlamydial growth. J. Med. Microbiol. 2003;52:409–415. doi: 10.1099/jmm.0.04992-0. [DOI] [PubMed] [Google Scholar]
- 19.Tormakangas L., Erkkila L., Korhonen T., et al. Effects of repeated Chlamydia pneumoniae inoculations on aortic lipid accumulation and inflammatory response in C57BL/6J mice. Infect Immun. 2005;73:6458–6466. doi: 10.1128/IAI.73.10.6458-6466.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aburatani S., Tashiro K., Savoie C. J., et al. Discovery of novel transcription control relationships with gene regulatory networks generated from multiple-disruption full genome expression libraries. DNA Res. 2003;10:1–8. doi: 10.1093/dnares/10.1.1. [DOI] [PubMed] [Google Scholar]
- 21.de Hoon M. J., Imoto S., Nolan J., et al. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 22.Mukhopadhyay S., Good D., Miller R. D., et al. Identification of Chlamydia pneumoniae proteins in the transition from reticulate to elementary body formation. Mol. Cell. Proteomics. 2006;5:2311–2318. doi: 10.1074/mcp.M600214-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Yu H. H., Di Russo E. G., Rounds M. A., et al. Mutational analysis of the promoter recognized by Chlamydia and Escherichia coli sigma(28) RNA polymerase. J. Bacteriol. 2006;188:5524–5531. doi: 10.1128/JB.00480-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen L., Feng X., Yuan Y., et al. Selective promoter recognition by chlamydial sigma28 holoenzyme. J. Bacteriol. 2006;188:7364–7377. doi: 10.1128/JB.01014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ide N., Ikebe T., Kutsukake K. Reevaluation of the promoter structure of the class 3 flagellar operons of Escherichia coli and Salmonella. Genes. Genet. Syst. 1999;74:113–116. doi: 10.1266/ggs.74.113. [DOI] [PubMed] [Google Scholar]
- 26.Mathews S., George C., Flegg C., et al. Differential expression of ompA, ompB, pyk, nlpD and Cpn0585 genes between normal and interferon-gamma treated cultures of Chlamydia pneumoniae. Microb. Pathog. 2001;30:337–345. doi: 10.1006/mpat.2000.0435. [DOI] [PubMed] [Google Scholar]
- 27.Mukhopadhyay S., Peiser L., Gordon S. Activation of murine macrophages by Neisseria meningitidis and IFN-gamma in vitro: distinct roles of class A scavenger and Toll-like pattern recognition receptors in selective modulation of surface phenotype. J. Leukoc. Biol. 2004;76:577–584. doi: 10.1189/jlb.0104014. [DOI] [PubMed] [Google Scholar]
- 28.Alper S., Duncan L., Losick R. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell. 1994;77:195–205. doi: 10.1016/0092-8674(94)90312-3. [DOI] [PubMed] [Google Scholar]
- 29.Zhong J., Douglas A. L., Hatch T. P. Characterization of integration host factor (IHF) binding upstream of the cysteine-rich protein operon (omcAB) promoter of Chlamydia trachomatis LGV serovar L2. Mol. Microbiol. 2001;41:451–462. doi: 10.1046/j.1365-2958.2001.02531.x. [DOI] [PubMed] [Google Scholar]
- 30.Fadel S., Eley A. Chlamydia trachomatis OmcB protein is a surface-exposed glycosaminoglycan-dependent adhesin. J. Med. Microbiol. 2007;56:15–22. doi: 10.1099/jmm.0.46801-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.