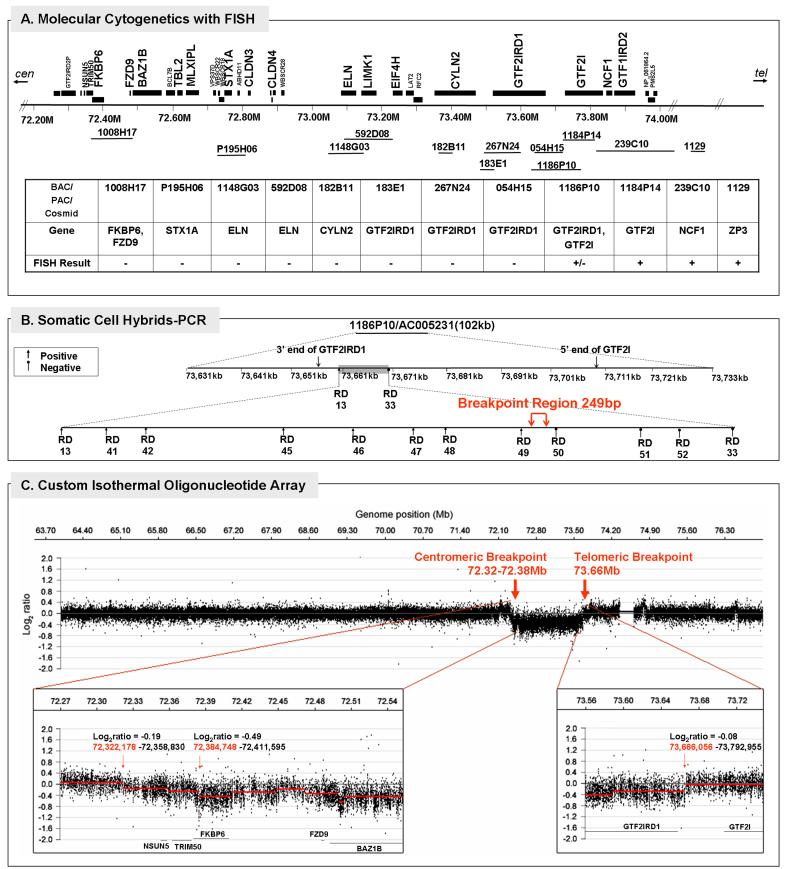
FIG. 2.
Deletion breakpoint analysis of Subject 5889. A: Physical map of BAC/PAC/cosmid probes used for molecular cytogenetics with fluorescence in situ hybridization (FISH) and summary of results defining deletion in 5889; B: Polymerase chain reaction (PCR) with somatic cell hybrids of the distal breakpoint region identified by FISH; C: High resolution array CGH analysis of a 14 megabase region on chromosome 7 (64-77Mb) using a custom isothermal oligonucleotide tiling array. The top panel shows the complete tiled region in a 10X window averaged view (200bp probe spacing). Bottom left and right panels show zoomed in views of the proximal and distal breakpoints for the deletion in which all data points (unaveraged data set with 385,013 probes and 20bp median spacing) were analyzed. Red line segments in both bottom panels denote copy number calls by the analysis algorithm (see Materials and Methods).