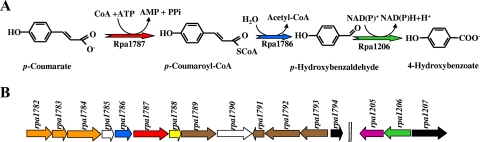
FIG. 10.
Proposed peripheral pathway for anaerobic catabolism of p-coumarate in R. palustris. (A) Enzymatic reactions of the pathway (267). The postulated enzymes are p-coumarate-CoA ligase (RPA1787), p-coumaroyl-CoA hydratase/lyase (RPA1786), and p-hydroxybenzaldehyde dehydrogenase (RPA1206). (B) Organization of the gene clusters likely to be involved in peripheral catabolism of p-coumarate in R. palustris CGA009 (GenBank accession number NC_005296). Genes are represented by arrows: red, RPA1787 gene, encoding p-coumarate-CoA ligase; blue, RPA1786 gene, encoding p-coumaroyl-CoA hydratase/lyase; green, RPA1206 gene, encoding p-hydroxybenzaldehyde dehydrogenase; orange and brown, genes encoding putative TRAP and ABC transporters, respectively; black, RPA1794 and RPA1207 genes, encoding putative transcriptional regulators of the MarR and Fis families, respectively; violet, gene encoding a putative aromatic alcohol dehydrogenase; yellow, gene encoding a putative thioesterase; white, genes of unknown function. Two vertical lines mean that the genes are not adjacent in the chromosome.