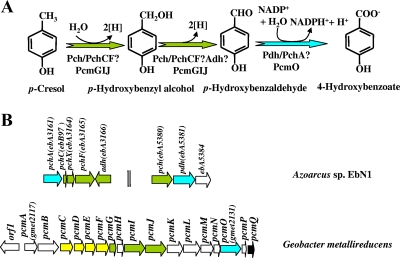
FIG. 13.
Proposed peripheral pathway for oxidation of p-cresol to 4-HBA in different bacteria. (A) Enzymatic reactions of the pathway in denitrifying (308) and iron-reducing (171, 277) bacteria. The enzymes are indicated as follows: Pch and PcmGIJ, p-cresol methylhydroxylase; Adh, p-hydroxybenzyl alcohol dehydrogenase; Pdh, PchA, and PcmO, p-hydroxybenzaldehyde dehydrogenase. (B) Organization of the gene clusters likely to be involved in anaerobic p-cresol oxidation in Azoarcus sp. strain EbN1 (GenBank accession number NC_006513) and G. metallireducens (accession number NC_007517). Genes are represented by arrows: green, genes known or predicted to encode p-cresol methylhydroxylase and a putative p-hydroxybenzyl alcohol dehydrogenase (adh); blue, genes predicted to encode a proposed p-hydroxybenzaldehyde dehydrogenase (or an aromatic aldehyde dehydrogenase); yellow, genes predicted to encode a membrane-bound cytochrome bc1 complex; white, genes of unknown function; black, the pcmQ regulatory gene of the p-hydroxybenzoate degradation cluster (Fig. 7). Two vertical lines mean that the genes are not adjacent in the genome.