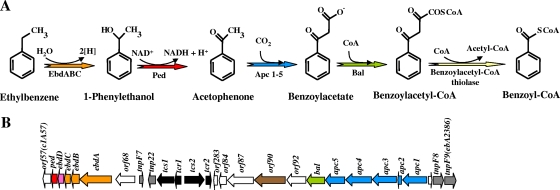
FIG. 17.
Proposed peripheral pathway for anaerobic catabolism of ethylbenzene in Azoarcus strains. (A) Enzymatic reactions of the pathway according to data described previously by Rabus et al. (292) and Johnson et al. (173). The enzymes shown are as follows: EbdABC, ethylbenzene dehydrogenase; Ped, (S)-1-phenylethanol dehydrogenase; Apc1 to Apc5, acetophenone carboxylase; Bal, benzoylacetate-CoA ligase. (B) Organization of the gene cluster involved in anaerobic catabolism of ethylbenzene in Azoarcus sp. strain EbN1 (GenBank accession number NC_006513) (203, 292). Genes are represented by arrows: orange, genes encoding ethylbenzene dehydrogenase; pink, gene encoding a putative ethylbenzene dehydrogenase-specific chaperone; red, gene encoding (S)-1-phenylethanol dehydrogenase; blue, genes predicted to encode acetophenone carboxylase; green, gene predicted to encode benzoylacetate-CoA ligase; brown, gene predicted to encode a transport system; black, genes encoding putative transcriptional regulators; gray, genes encoding putative transposases or transposase fragments; white, genes of unknown function.