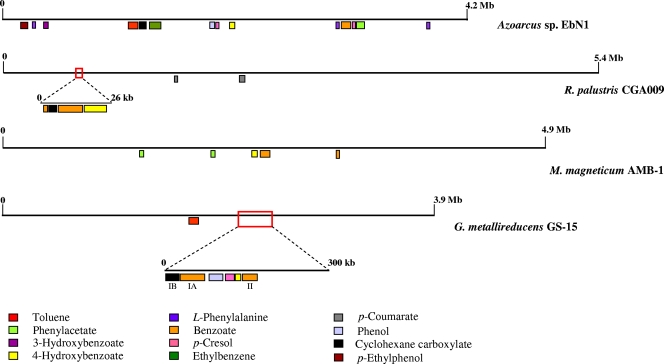
FIG. 18.
Comparative distribution of the gene clusters involved in anaerobic catabolism of aromatic compounds in different bacterial chromosomes. Characterized or predicted gene clusters for the anaerobic degradation of different aromatic (and cyclohexane carboxylate) compounds are indicated with a different color code. The supraoperonic clustering (26 kb) and the catabolic island (300 kb) of R. palustris (RPA0650 to RPA0673) and G. metallireducens (gmet2037 to gmet2284), respectively, are boxed and expanded. The gene arrangements within the clusters are shown in detail in Fig. 2 to 17.