Abstract
Summary: Mutants of translation components which compensate for both −1 and +1 frameshift mutations showed the first evidence for framing malleability. Those compensatory mutants isolated in bacteria and yeast with altered tRNA or protein factors are reviewed here and are considered to primarily cause altered P-site realignment and not altered translocation. Though the first sequenced tRNA mutant which suppressed a +1 frameshift mutation had an extra base in its anticodon loop and led to a textbook “yardstick” model in which the number of anticodon bases determines codon size, this model has long been discounted, although not by all. Accordingly, the reviewed data suggest that reading frame maintenance and translocation are two distinct features of the ribosome. None of the −1 tRNA suppressors have anticodon loops with fewer than the standard seven nucleotides. Many of the tRNA mutants potentially affect tRNA bending and/or stability and can be used for functional assays, and one has the conserved C74 of the 3′ CCA substituted. The effect of tRNA modification deficiencies on framing has been particularly informative. The properties of some mutants suggest the use of alternative tRNA anticodon loop stack conformations by individual tRNAs in one translation cycle. The mutant proteins range from defective release factors with delayed decoding of A-site stop codons facilitating P-site frameshifting to altered EF-Tu/EF1α to mutant ribosomal large- and small-subunit proteins L9 and S9. Their study is revealing how mRNA slippage is restrained except where it is programmed to occur and be utilized.
INTRODUCTION
Relative ratcheting movement of ribosomal subunits and swiveling of the head domain are fundamental features of the ribosome cycle, likely tied to progressive triplet mRNA movement. Even without any ribosome structural knowledge, it would be easy to imagine that the complexity of decoding is such that any one mutant, or a simple combination of mutants, could “break” the machine but not alter it so that at a detectable rate, the mRNA would be moved by a net two or four, rather than three, nucleotides. Indeed, at the time that the code was being deciphered, it was thought that triplet decoding would be immutable. At a later stage Crick described his congruent views and observations at the time (67). One consequence of immutability would be that mutants of translation components which would allow compensatory frameshifting near the site of a frameshift mutation (indel, an insertion or a deletion), and so restoration of some ribosomes to the wild-type (WT) reading frame, could not be found. Such a lack of external suppressors was even used in 1966 as a criterion for a mutant to be considered a candidate frameshift mutant (340). Another criterion was a lack of even a low level of functional product, i.e., leakiness, due to detectable compensatory error frameshifting near the indel site by WT translation components (340) (At this stage the ability to sequence DNA was almost a decade in the future [203, 276]). However, mutations which were shown to be frameshifts were found to be externally suppressible (two “wrongs” making a partial “right”) (263, 266, 357). In addition, leakiness of frameshift mutants was detected. The prelude to one study was isolating bacterial mutants with a frameshift mutation-inducing mutagen, discarding the leaky mutants, and studying the remainder as candidate frameshift mutants (223). Subsequently these frameshift mutants, of both signs, were shown to be leaky, even though the degree of leakiness was by then no longer representative of an unbiased sample (10). However, the finding of notable levels of frameshifting at or near a run of Us (98) reinforced the point. The finding of −1 frameshifting at the central codon of the sequence UUA AAG GGA (but not detectably at its counterpart WT sequence UUA CAG GGA) further indicated the identity of a shift-prone site (14). Other early work revealed −1 frameshifting on alteration of the balance of aminoacylated tRNAs (12, 106, 331). Part of these latter studies involved manipulation of the level of aminoacylation. Since severe nutrient limitation is common in nature, frameshifting caused by aminoacyl-tRNA limitation may be utilized in gene expression. The other study involved relative tRNA concentrations. A main motivation in this case was relevance of the frameshifting involved to the synthesis of a phage-encoded product naturally synthesized under replete conditions (12). However, possible functional significance was not pursued, and though a different type of phage was shown shortly afterwards, in 1983, to also encode frameshift products (89), the nature of the frameshifting involved was not followed up until much later. In 1984 machines for the routine synthesis of oligonucleotides of specified sequence became available, and shortly afterwards, the field underwent a major change.
The breakthrough in 1985 to 1987 was the discovery of cases where without amino acid starvation, frameshifting plays an essential role in gene expression (62, 64, 135, 151, 152, 205, 333). In addition to the frameshift-prone site, these cases generally involve signals, often called recoding signals (111), embedded in the mRNA that greatly stimulate the level of frameshifting at the shift site. There are codes within the code, and redirection of linear readout is an important component of the reprogramming of decoding, or recoding (23, 92, 110, 219). Such programmed frameshifting will not be included in this review, except for the nature of shifting at a few of the shift-prone sites in the absence of recoding signals.
Instead, this review will focus on how ribosomes can shift frame at simple sequences and the implications of this for standard reading frame maintenance. Can the fundamental ratcheting mechanism be altered so that its relevant part does not move the distance required for triplet translocation? If not, can the ribosome loose the grip it must have on either or both parts of the tRNA-mRNA paired complex in a way that permits reading frame realignment? Alternatively, at the end of a ratcheting cycle can there be dissociation of anticodon-mRNA pairing which permits realignment prior to their re-pairing? Pertinent to possible relevant alterations of the ratcheting mechanism are studies on mutants of rRNA that cause frameshifting or influence programmed frameshifting efficiency and on the mechanism of action of recoding signals. Salmonella and Escherichia coli each have seven copies of their rRNA genes, and Saccharomyces cerevisiae has even more. All the initial studies with revertants of frameshift mutations in these organisms used strains with the WT complement of their rRNA genes. Therefore, in these initial studies only dominant mutants would have been recovered. Even though later studies rectified this, that work will not be included here as it is being prepared for review elsewhere (J. D. Dinman and M. O'Connor; personal communication).
Even though the events involved in the specification of any amino acid are a continuous process, the characteristics of three ribosomal sites, A, P, and E, are central to an understanding of the redirection of reading frame and also of frame maintenance. Obviously, the relatively recent breakthroughs in atomic-level structural knowledge of bacterial ribosomes (Fig. 1 to 3), at least in certain conformations, are of prime importance. Further development of this knowledge will set the stage for an understanding of the relative functional importance revealed by genetic studies. Although it was a stroke of genius to postulate an adaptor before tRNA was discovered (66), it is now apparent that tRNA is far from a passive adaptor. Not only does the angle between its arms change during translation, but there is a contrast between the tightly stacked and organized anticodon loop in the A site and the widened feature of its P-site counterpart (282, 300; reviewed in reference 170).
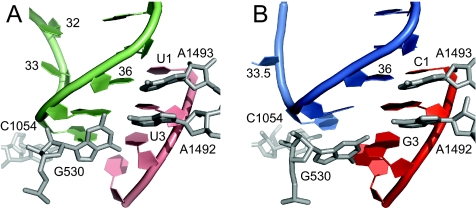
FIG. 1.
The anticodon-codon interaction in the A sites of a tRNA with a normal three-nucleotide anticodon (A) and a tRNA with a four-nucleotide anticodon (B) in the same site. (Panels B and D were modified from reference 88 with permission of the publisher.)
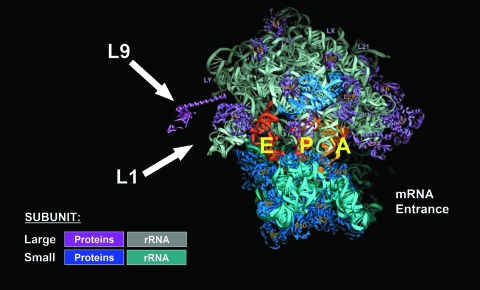
FIG. 3.
Structure of the 70S ribosome. Positions of proteins L1 and L9 are indicated with arrows. The cleft where mRNA entering occurs is also shown. (Modified from reference 358 with permission of AAAS.)
Even with some of the relevant WT conformations in mind, the need for caution in making inferences from mutants without structural information about them is obvious. An illustration of this comes from X-ray crystallographic analysis of an anticodon loop with eight rather than the standard seven nucleotides. It has an anticodon with the standard number of three nucleotides, but they span four codon bases (Fig. 1A and B) (88). This is obviously mechanistically relevant, especially because much of the early discussion of +1, but not −1, frameshift mutant suppressors focused on the inference that anticodon size determines codon size. This first foray into high-resolution pertinent ribosome structural knowledge by Dunham and Ramakrishnan and their colleagues is a great advance for the field.
The productive mixture of genetic and biochemical analyses makes some terminology clarification desirable. Abbreviations for modified nucleosides are as described by Limbach et al. (189), and an updated version can be found at the website http://medstat.med.utah.edu/RNAmods/. A compilation can also be found in reference 213. Subscript and a superscript numbers indicate the number and the position of a substitution, respectively; e.g., 6-dimethyladenosine is abbreviated m62A. c-, i-, k-, m-, n-, o-, r-, s-, and t- are abbreviations for carbonyl, isopentenyl, lysyl, methyl, amino, oxy, ribosyl, thio, and threonyl groups, respectively. An abbreviation to the left or to the right of the nucleoside symbol denotes a modification of the base or the ribose, respectively. Other nucleoside abbreviations are as follows: T, m5U or ribothymidine; Ψ, pseudouridine; I, inosine; Q, queuosine; R, purine; Y, pyrimidine; and N, any of the four major nucleosides. A number following an abbreviation for a modified nucleoside denotes the location in the tRNA sequence. An enzyme catalyzing the formation of, e.g., Ψ at position 38 is denoted tRNA(Ψ38) synthetase and likewise for other modifying enzymes. tRNA species are identified by their anticodon sequence. N34 denotes the nucleoside in position 34 (wobble position) of the tRNA, and N(III) denotes the third nucleoside of the codon. ASL indicates the anticodon-stem-loop domain of a tRNA usually consisting of 17 nucleotides.
When four mRNA bases are decoded as a single amino acid, the term quadruplet decoding is sometimes used in the genetic literature. In this sense, “decoding” refers to the overall end result. In contrast, many biochemists use “decoding” to mean specifically events in the ribosomal A site. With the latter usage, quadruplet decoding would classically mean quadruplet Watson-Crick base pairing in the A site. The key issue is whether quadruplet decoding is interpreted to imply quadruplet translocation or not. From the overall end-result usage, the increased frameshifting efficiency often found when there is the potential for Watson-Crick pairing of the four mRNA bases (or wobble pairing of the fourth) can be solely due to events in the ribosomal P site (whether all four potential anticodon bases are simultaneously involved in Watson-Crick pairing with four codon bases or whether the first three pair and the first one dissociates before the fourth pairs to mRNA is a secondary issue). With this scenario, three-mRNA-base translocation and subsequent mRNA slippage can be involved. Because of potential misunderstanding as to what is implied by doublet or quadruplet decoding, we will not use these terms. Similarly, the term “near cognate” as applied to tRNA is potentially ambiguous in the present setting. Instead we will use the term “third-position mismatched,” and associated definitions follow are as follows. (i) “Third-position mismatched” refers to cases with Watson-Crick pairing in positions 1 and 2 but where there is not Watson-Crick or G34-U wobble pairing in position 3. In many but not all cases, third-position mismatched will be the same as near cognate; e.g., proM tRNAcmo5UGGPro is third-position mismatched according to this definition and also near cognate, interacting with the C-ending codon. I34-A(III) is third-position mismatched, since it has a very weak interaction with A (78). (ii) “Cognate” refers to cases with Watson-Crick pairing in positions 1, 2, and 3 or Watson-Crick pairing in positions 1 and 2 and a G34-U wobble in position 3. The interaction of I34 with U- and C-ending codons is cognate, since it makes a normal Watson-Crick base pair with these bases, but I34-A is considered third-position mismatched (see above). (iii) “Noncognate” refers to cases with mismatches in position 1 or 2.
These are the operative definitions used here. Depending on the progress of research, a base derivative may pair in a well-defined manner. An example of this is the coding capacity of N2-lysylcytidine (k2C) (also denoted lysidine), which, according to the operative definition, is third-position mismatched, but we know now that the modification completely changes the coding capacity of the wobble C to base pair with A in a Watson-Crick configuration.
The great majority of base substitution mutations in coding sequences, and not just in third-codon positions, have little if any obvious phenotypic effects. However, shifting reading frame often makes a gibberish product from the new frame (with important exceptions being in utilized programmed frameshifting). While selection has operated to keep wasteful frameshifting at a low level, it has not been at a minimal level, arguably because of speed considerations. Nevertheless, it has been proposed that the advantage of keeping nonprogrammed frameshifting low has been sufficiently strong to drive selection of a third ribosomal site, the E site. The proposal is that the E site restrains mRNA slippage by ensuring that the anticodons of two, and not just one, tRNAs are always bound to mRNA, by requiring that E-site deacylated tRNA does not lose its grip on mRNA before an A-site tRNA anticodon has commenced pairing with mRNA (reviewed in reference 345). This proposal is highly attractive, and though it is controversial, there are increasing genetic data for it (see below). Regardless of its merits, key aspects of translocation remain to be resolved.
Future developments in atomic-resolution crystallographic structures, cryoelectron microscopy-generated conformational insights coupled with fluorescent resonance energy transfer analyses, and rapid kinetic measurements will provide the framework necessary for an understanding of the still-elusive mechanism of framing. While the structures and their conformational changes during the ribosome cycle are crucial, functional significance, as revealed by mutant analysis, is relevant as well. The present review brings together the disparate studies of tRNA mutants relevant to framing. As most of these tRNA mutants were isolated before ribosome structural knowledge became available and detailed questions meaningfully posed, their degree of usefulness is expected to vary and is to some extent unknown at present. Most of these mutants were isolated as suppressors of frameshift mutations in biosynthetic genes, but some, especially the modification mutants, were isolated previously and later tested for their ability to suppress frameshift mutations (some suppressors which are not tRNA mutants are also included). The finding of suppressors paved the way for the discovery of utilized natural frameshifting. While studies of extragenic suppressors of frameshift mutants started much later in the 1960s than those of nonsense mutants, an understanding of their mechanism of action has been actively pursued for much longer. Their study has generated the diverse insights to be presented here, many of which were not apparent at the time of a previous review (16).
ORIGIN AND IDENTITY OF EXTRAGENIC FRAMESHIFT MUTANT SUPPRESSORS
The Classic Set of Suppressors Isolated as Revertants of Salmonella Histidine Operon +1 Frameshift Mutations
There were two seminal papers in 1970 on mutation-induced +1 frameshifting. Yourno and Tanemura identified the amino acid specified by four bases in a strain with an external suppressor of a +1 frameshift mutation which acted at that site (357). Riddle and Roth selected various extragenic suppressors (sufA, -B, -C, -D, -E, and -F) to different frameshift mutations in the his operon (263) which were subsequently shown to have single-nucleotide insertions, i.e., +1. These suppressor mutants have been important tools in several investigations aiming to clarify the mechanism of reading frame maintenance. Although many of them were thoroughly analyzed shortly after their original isolation (264, 265), subsequent investigation of some of them has revealed new aspects of how frameshifting can occur.
The molecule suspected of causing the frameshift mutant suppression was tRNA, and indeed it was demonstrated early for sufA and sufB mutants that one of the three proline tRNA species had an altered chromatographic migration (265). The frameshift product had proline at the site corresponding to the suspected frameshift site, which was CCC-UGA in the hisD3018 +1 frameshift mutant (357). However, only much later did it become known that the sufA6 mutation is an insertion in the anticodon of tRNACGGPro (encoded by the proK gene) (184) and that the sufB2 mutant has an insertion in tRNAGGGPro (encoded by the proL gene) (295).
The sufC mutants have suppressor specificity overlapping that of sufA and sufB, but unlike these suppressors, sufC was suggested to be recessive (264, 275). Since sufC has the same suppressor specificity as sufA6 and sufB2, sufC might induce a modification deficiency of the proline tRNAs affected by the sufA6 and sufB2 mutations. However, it later emerged that the three original sufC mutants (sufC10, -13, and -14) each contain mutations in two genes, denoted sufX and sufY (295). The sufX mutations are in the proL gene and thus are allelic to sufB2. One of these sufX mutations (sufX201) is a base substitution (G43A [Fig. 4 shows tRNA numbering]) at the junction between the anticodon stem and the TΨC stem in the proL tRNAGGGPro (295). This alteration in tRNAGGGPro, as well as many other base substitutions and base insertions such as sufB2, induces frameshifting by the third-position-mismatched proM tRNAcmo5UGGPro (258, 259).
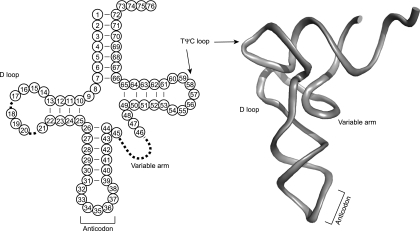
FIG. 4.
(Left) Cloverleaf structure of a standard tRNA with the conventional numbering system for the locations of the different nucleotides used in the text. (Right) Three-dimensional structure of yeast tRNAPhe with the various regions of the tRNA indicated.
In addition to a sufX mutation, each of the three original sufC mutants also contains a mutation (sufY) that induces +1 frameshifting (295). The three sufY mutations isolated in 1970 are dominant (“gain of function”), and they induce an amino acid substitution at the same position (position 67) of the YbbB protein (60). This protein, which contains a rhodanese domain, is required for the in vitro exchange of sulfur of mnm5s2U34 with selenium, forming mnm5Se2U34 in tRNA (350). The altered SufY (YbbB) protein showed a novel activity, since tRNA from the mutant has a previously unknown modified nucleoside in the wobble position of the anticodons of Lys-, Glu-, and Gln-tRNAs. The novel nucleoside has a C10H17 fragment (most likely a geranyl group) added to the s2-group of wobble nucleoside mnm5s2U34 in these tRNAs (60). How this mediates compensatory frameshifting to suppress frameshift mutations is considered below.
The sufD42 mutant was the first frameshift suppressor mutant to be sequenced. Riddle and Carbon showed that it has an extra C in the anticodon loop of the major tRNACCCGly, creating a possible four-base anticodon (262). This followed earlier work showing that the suppressor is genetically dominant and that in mutant cells glycine tRNA has changed chromatographic behavior (264, 265). Its existence led to a quadruplet-translocation hypothesis, which is considered in detail below.
The sufE mutations, which were induced by a frameshift mutagen, are dominant and are located close to the thi gene (263, 264). Their specificity suggested that these suppressors might contain an altered Gly-tRNA since they suppress his alleles which are also suppressed by sufD42 (an allele of glyU encoding tRNACCCGly). However, we know now that close to the thi locus there is an operon containing four tRNA genes. One of these, glyT, specifies tRNAmnm5UCCGly, which decodes both GGA and GGG. Moreover, it has since become known that hisC3072, hisC3736, and hisD3068, which are suppressed by sufE, contain in their frameshift windows the mRNA sequences -CCG-GGG-GAA, -CAG-GGG-AUU-, and UAU-GGG-GCC-, respectively (bold indicates the inserted nucleoside, and spacing denotes the zero frame) (180). Thus, these sequences have a possible frameshifting site for a GGG-decoding Gly-tRNA, consistent with sufE mutations altering tRNAmnm5UCCGly.
The sufF44 mutation is recessive and imposes changes in the chromatographic behavior of tRNAmnm5UCCGly (also known as tRNA2Gly). Since the sufF44 mutation was not located in an area of the chromosome where any of the glycine tRNA genes are located, the sufF gene was suggested to encode a tRNA modification enzyme (264, 265). However, it was later shown to be mutated in the argU gene, which codes for tRNAmnm5UCUArg (179). The alterations in tRNAmnm5UCUArg cause instability or less-efficient arginylation, resulting in a decreased concentration of charged tRNAmnm5UCUArg. Presumably this results in slow decoding of its cognate A-site codon, which, when there is a 5′ adjacent CAA codon, facilitates +1 slippage by peptidyl-tRNAcmnm5s2UUGGln, thereby restoring some translation to the WT reading frame. (The changed chromatographic behavior of tRNAmnm5UCCGly was suggested to be due to excessive frameshifting in either of the two genes gidA and mnmE, which are required for the synthesis of the mnm5 side chain, even though the great majority of the products should still be WT. This suggestion is even less tenable because the modification pattern of tRNAmnm5UCCGly from the sufF mutant was found to be indistinguishable from WT [179]. A possible caveat remains; as the mnm5U was not chromatographically separated, the changed chromatographic behavior of tRNAmnm5UCCGly may still be due to lack of the mnm5 group of its wobble nucleoside. However, the codon AGA, which is read by tRNAmnm5UCUArg and is defective in sufF mutants, is used only once in decoding gidA [though it is in the shift-prone sequence UUU-AGA {105}] and not at all in mnmE mRNA. We consider the possibility that there is an alteration of tRNAmnm5UCCGly which is relevant to the frameshifting to be very low.)
In 1983 Roth and colleagues reported the isolation of additional frameshift mutant suppressors (sufG70, sufH90, sufI91, sufJ128, and sufM95) for various frameshift mutations in the his operon (166). The sufH90 and sufI91 mutations were mapped at different locations, but they were very unstable and not much work could be done with them. The sufM95 suppressor has a suppression pattern distinct from that of any of the earlier-isolated suppressors, including sufA6, yet it maps to the sufA locus. It still may be allelic to sufA6 and thus a derivative of tRNACGGPro. sufJ128 has an extra nucleotide in the 5′ part of the anticodon loop, potentially creating an anticodon of four nucleotides (discussed below). The sufG suppressor was thought to suppress at runs of A and therefore likely to be a mutated tRNAmnm5s2UUULys (167). However, it was later shown to be a derivative of tRNAcmnm5s2UUGGln and to suppress at CAAA sequences (228).
Establishing that the First Suppressible Candidate Frameshift Mutants Were Really Frameshift Mutants: Suppressors for Frameshift Mutations of Both Signs
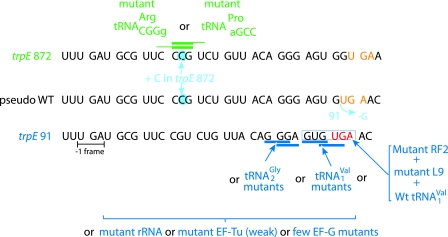
Before the advent of DNA sequencing, a primary concern in finding extragenic suppressors of a mutant classified as a frameshift mutant was that the classification was correct. With Salmonella enterica hisD gene mutants used to select the suppressors just described, this was solved by genetically isolating revertants with nearby compensatory frameshift mutations (intragenic revertants due to −/+ or +/− combinations) and sequencing the relevant peptide(s) from the encoded product (354-357). With an S. enterica anthranilate synthetase gene mutant, corresponding revertants due to intragenic compensatory mutations were previously isolated. However, in this case the individual frameshift mutations were separated into different strains. This was accomplished by transductions. The polarity and mutagenic properties of the isolated mutants could be explicable only by their being frameshift mutants (the starting mutant was at the time known as S. typhimurium tryA91 [266] but with subsequent nomenclature changes is now called S. enterica trpE91) (Fig. 5). Years later, DNA sequencing confirmed the hint from mutagenesis data that trpE91 itself was a −1 rather than a +1 mutation (14). Its first external suppressors were reported in 1968 (266). The trpE872 frameshift mutant, which was itself isolated as an internal suppressor for trpE91, is also externally suppressible (15), so it was obvious from an early stage that frameshift mutants of both signs are externally suppressible. However, it was not until after trpE872 was shown by DNA sequencing to be a +1 frameshift mutant (14) that its selected suppressor was characterized in detail and suppressors of a different class constructed (317) (Fig. 5). The original suppressor, sufT621, had an extra base, G, between bases 36 and 37 in a gene for tRNA2Arg, resulting in an anticodon complementary to CCG(U) (whether the anticodon is 5′ ACGg or 5′ ICGg, where I is inosine, is unknown). Suppression also occurs with mutants with 7- and 9-membered anticodon loops as well as when a tRNAPro has an extra base, A, and a predicted anticodon 5′ ACGG (and another change) (316, 317). Curiously, one of the suppressors with two extra nucleotides in its anticodon loop also has 10 extra bases in its TΨC arm (316).
FIG. 5.
Suppressors of a −1 frameshift mutation and of a nearby +1 mutation. S. enterica trpE91 has a deletion of G400 in its anthranilate synthetase gene (blue), which if not compensated for leads to termination at a UGA codon (brown). A fast-growing (pseudo-wild) revertant on media without tryptophan had an insertion of a C at 18 nucleotides 5′ (blue). The secondary compensatory mutation was separated by transduction and designated trpE872. The anticodons of tRNA mutants which suppress trpE872 are in green. One derivative of tRNAmnm5UCCGly (tRNA2Gly) directly suppresses trpE91 by −1 frameshifting at G GGA, and others do so indirectly. An anticodon insertion mutant of tRNAcmo5UACVal causes slipping +2 from GUG to GUG, and another mutant has C74 substituted. Mutants of L9 and RF2 facilitate +2 slippage by WT tRNAcmo5UACVal from GUG to GUG.
Diverse Suppressors of a Salmonella −1 Frameshift Mutant: Action via −1 or +2 Frameshifting
As expected from the analysis of phage T4 rII mutants (25, 68), the growth of almost all intragenic revertants (+/− combinations) of the −1 frameshift mutant trpE91 was nearly as fast as that of the WT on media lacking tryptophan. However, intragenic suppressors were sought among the very slowest growing revertants, and two were identified. Though independently isolated, their secondary mutation was identical and DNA sequencing confirmed an inference from mutagen studies that this was a base substitution mutation rather than a nearby +1 frameshift mutation (14). The substitution, C to A, changes the sequence just 5′ of the site of the frameshift mutation from UUA CAG GGA to UUA AAG GGA. The favored explanation at the time was that the natural compensatory frameshift involves a −1 shift to AAA from AAG. This was strongly supported by the finding that A AAG, especially as part of the sequence A AAA AAG, is highly shift prone in E. coli (314, 335). (The reason for the −1 shift-prone nature of A AAG is described below.) (Some of the compensatory trpE frameshift mutants isolated from revertants of trpE91 in the [+ or −] combination splitting experiment described above were leaky, i.e., capable of slow growth without added tryptophan and without any secondary mutation. However, whether a particular compensatory frameshift mutant exhibited leakiness or not was simply explicable [14]. Interestingly, compensatory frameshifting at a WT shift-prone site is important for reactivation of several acyclovir-resistant mutants of herpes simplex virus and is a medical issue [35, 115].)
The initial set of trpE91 external suppressors comprised two classes (Fig. 5). The strongest of these, e.g., sup-601, and several of intermediate strength, e.g., sup-617, were later renamed sufS601, sufS617, etc., and are alleles of the glyT, gene which encodes tRNAmnm5UCCGly (tRNA2Gly, as described below). sufS601 does not act as a suppressor of UAG, UAA, or UGA (i.e., resulting in any of these triplets specifying an amino acid) and did not suppress any of the eight presumptive his frameshift mutants tested in the initial study (or any of the 69 his frameshift mutants tested later) (245). However, the very weakest of the initial set of trpE91 external suppressors caused more efficient (triplet) readthrough of a UGA stop codon (UGA was reported to be a stop codon in 1966, and UGA mutants of Salmonella enterica became available only as the trpE91 suppressor work was first being submitted the following year [34, 271]). The most efficient of this class of suppressors were not viable on nutrient agar or other rich media and grew only on minimal media, permitting selective transductions to be performed on rich media. They were later shown to be alleles of the supK gene, described in the intervening period (260). In 1988, it was shown that supK encodes release factor 2 (RF2), which mediates release at UGA and UAA, and it become known as prfB (159). However, earlier it was thought that the supK gene encoded the enzyme [tRNA(mcmo5U34)methyltransferase] catalyzing the formation of the methyl-ester of uridine-5-oxyacetic of wobble position of certain tRNAs (254, 255, 261), and this modification has important coding consequences (220). The reason for a reduced activity of the tRNA(mcmo5U34)methyltransferase in the supK mutant has not been clarified, but it might be due to a secondary effect of the defective RF2 on the formation of this enzyme. Interestingly, there is a UGA stop codon between the cmoA and the cmoB genes, which encode enzymes involved in the synthesis of mcmo5U34 (220). Increased readthrough of this UGA stop codon might interfere with the synthesis of this enzyme.
An understanding of how UGA suppressors, which, it emerged, were due to defects in RF2, mediated suppression of the −1 frameshift mutant trpE91 showed that two early suggestions were incorrect (one was based on the distraction provided by the reported methylase defect of supK mutants described in the last paragraph) (15, 266). The 3′ sequence flanking the site of the G deletion in trpE91, compared to WT, is GGA GUG UGA. Extra-slow decoding of UGA due to defective RF2 is thought to increase the chance for GUG-decoding WT tRNAVal in the ribosomal P site to detach from pairing to the zero-frame GUG and to re-pair to mRNA at the underlined +2 GUG. This returns the reading to the original frame in the WT (GGA GUG GUG A) with the omission of one amino acid (C. Johnston et al., unpublished data). The strain also had a mutant ribosomal protein L9 gene (Johnston et al., unpublished data cited in reference 17), which perturbs the role of L9 in restraining forward mRNA slippage (1, 128, 130, 131) (defects in L9 are also known to suppress the +1 frameshift mutant hisC3072 [180] [see below and Fig. 3]). While it is not known how the trpE91-containing strain came to have two suppressor mutations, it is likely relevant that trpE91 is very weakly suppressed by mutant L9 on its own at 22°C even though this is not detectable at 37°C (M. O'Connor, personal communication). Also, even though otherwise WT, trpE91-containing strains are nonleaky on minimal media without tryptophan, when they are supplemented with all other amino acids except for tryptophan, growth is detectable. Presumably a mutation giving one of the two suppressors allows a sufficient increase in cell numbers so that the chance of a second enhancing suppressor is increased (as noted above, the classic sufC suppressor also contained two mutations).
A different and more efficient class of trpE91 suppressors achieved the same +2 GUG UGA reading and involved only a single mutation. These suppressors, hopR, were characterized molecularly (235) before trpE91 suppression by defective RF2 mutants was understood, and they yielded provocative deductions about alternate anticodon loop stacking (see below). Though the initial hopR mutants had an extra base in their anticodon loops (235), later mutants with base substitutions in the anticodon stem and a base deletion in the variable loop (230) illustrated the variety of alterations in tRNAVal which also can cause frameshifting.
Another class of tRNAVal mutants isolated as suppressors of the −1 frameshift trpE91 were A or G substitutions of C74, the 5′-most C of the universally conserved CCA at the 3′ ends of tRNAs (239). These are described in detail below.
Other external suppressors of trpE91 were in the genes for elongation factor EF-Tu (142, 318), EF-G (193), and especially rRNA genes (231-234, 236-238, 277, 310). The former two are described below, and the latter will be reviewed elsewhere by J. D. Dinman and M. O'Connor. As described below, even overexpression of WT tRNA1Gly resulted in suppression of trpE91 (227).
Despite the amount of work performed on revertants of trpE91, it is likely that other classes could be found, especially under conditions where the common classes are precluded. While such a search is now unlikely, mutants selected as revertants of other frameshift mutants, e.g., the suppressors with mutant ribosomal protein S9 or constructed rRNA mutants, may be tested for their ability to suppress the −1 frameshift mutant trpE91. What is now known as trpE91 was generated by X-ray mutagenesis in the mid-1950s in Cold Spring Harbor Laboratory and first provisionally characterized as a frameshift mutant by Bauerle and Margolin (27). Since then, the sequence GGA GUG UGA at the 3′ end of its short frameshift window has proved useful in demonstrating the latitude, rather than the imperviousness, of mutants of translational components to mediating frameshifting.
Suppressors of +1 Frameshift Mutations in the S. cerevisiae HIS4 Gene Show Preferences and Diversity
Though the investigation of frameshift mutant suppressors in yeast (71, 86) started significantly later than the investigation of those in bacteria, its thoroughness was exemplary (70, 73). Like their counterparts in Salmonella utilized by J. Roth and colleagues, the characteristics of the S. cerevisiae his +1 frameshift mutants used for these studies (199) reflected the mutagen used for their isolation, the acridine derivative ICR191 (4). The resulting mutations are predominately in runs of Gs or Cs, and though suppressors are commonly not restricted to act at the mutation site, most of the initial yeast set were mutants of glycine or proline tRNAs. Several directly involved a base insertion into the anticodon loop (76, 206), whereas another had a base substitution in the anticodon stem (103) which appeared to result in an enlarged anticodon loop (75). M. Culbertson and colleagues reached two important conclusions from the study of these mutants, and they are described below.
Several of the yeast frameshift mutant suppressors were not tRNA mutants (72). One class of these were mutants of EF-1α (272) (those mutants and their bacterial counterparts are considered below). Another class were alleles of a gene, SUF12, which has regions of homology with EF-1α (346) but is quite distinct. Alleles of this gene have been isolated independently in several different selections (85). The gene is now known as SUP35, and it encodes eRF3 (360), for which PSI is a prion form (218, 309, 341). Yet other of the yeast frameshift mutant suppressors were in genes for a transcription factor and also ribosomal protein S3 (127).
From the Early Studies to Evolving Concepts of tRNA Mutant-Mediated Frameshifting: the Yardstick Model
Early studies of frameshift mutant suppressors did not address whether the frameshift event occurred in the A or the P site, and the authors interpreted their data cautiously. Hardesty et al. (123) suggested a P-site slippage model, and Yourno and Tanemura (357) also considered slippage as one of their two possible models to explain how +1 frameshifting occurs. Presuming that the anticodon was expanded by one base, they alternatively suggested that four-nucleotide base pairing and thereby a quadruplet translocation occurred. The latter model became the prevailing view and was strongly reinforced by the finding that the sufD suppressor had an extra base in the anticodon (262). The appealing concept was generalized to suggest that the tRNA anticodon governs the length of the translocation step—the yardstick model. It was adopted in several textbooks published during 1970s and 1980s (183, 294, 301, 330) (e.g., on page 102 in reference 183, it is stated, “The distance of three bases that the ribosome moves is probably determined by the interaction between the codon on mRNA and the anticodon on tRNA”). Attractive as the yardstick model was, it has been questioned (172) and has been proven invalid for two frameshift suppressor derivatives of Pro-tRNAs (sufA6 and sufB2) having an extra nucleotide in the anticodon loop (259). The yardstick model received much less attention from those studying frameshifting by WT tRNAs, especially programmed frameshifting, but nearly all of this area developed much later than the yardstick model originated.
In studies with frameshift mutant leakiness (10, 14, 98), tRNA balance (12, 106), and programmed frameshifting during the 1980s, it was assumed or shown that WT tRNAs with standard seven-nucleotide anticodon loops mediated the frameshifting involved. Also, in at least some cases it was assumed that the frameshifting occurs in the P site (e.g., for the RF2 programmed frameshifting [64, 333] and translational hops [333]), and this was explicitly stated in the case of Ty1 transposition by Belcourt and Farabaugh (28) and by Qian and Björk (258). To explain the frameshifting involved in these cases, and also tandem −1 frameshifting, there seemed no realistic alternative to slippage of the mRNA relative to the peptidyl-tRNA anticodon. The investigators involved did not address the yardstick model, as it was not considered a relevant issue. However, even during the present decade, in several reports of tRNAs with enlarged anticodon loops, the mechanism of quadruplet translocation is still invoked (see, e.g., references 5 and 194). It was suggested that within limits, tRNA acts as a “molecular ruler” to determine the codon size during translation of the mRNA (5, 306). These studies involve the construction of tRNAs with an extra base in their anticodon loops as part of schemes to expand the genetic code lexicon. This topic is considered in detail below, but its essence is that synthetic expansion of the number of types of amino acids encoded requires extra codons to encode the novel amino acids. Having tRNAs acylated with the novel amino acids preferentially decode specific quadruplet codons featured as one of the approaches tested. Recent direct investigations of how a tRNA with an expanded anticodon loop induces frameshifting were interpreted as supporting quadruplet translocation in these cases (253, 326).
Though it is not explicitly stated, for some the yardstick model is still influential. Certain others consider that anticodon size does not in general determine codon size but that when the potential for four-base codon-anticodon interaction occurs in the A site, translocation can be directly quadruplet. The rest consider that regardless of the type of codon-anticodon which is accepted in the A site, translocation is invariably triplet but mRNA-anticodon realignment can subsequently occur and mediate alternative framing. The present perspective deals with these possibilities and demonstrates how an attractive model consistent with several experimental results can be self-perpetuating and even incorporated into several textbooks despite the undermining effect of the parts of the data discussed below which were available at the time.
CONTRASTING P- AND A-SITE RIBOSOMAL ENVIRONMENTS: MODE OF TRANSLOCATION BETWEEN THEM
The ribosomal P site functions to hold the peptidyl-tRNA in a secure grip in order to position it for peptidyl transfer. It likely also serves to maintain the reading frame. Evidence for the latter can be inferred from the studies of programmed frameshifting and the mutant studies reviewed here. It is an essential starting point for the proposal that two tRNAs are always paired to mRNA to facilitate frame maintenance (345). Data from other sources also are consistent with or support such a role, although experimental results to support the latter suggestion are scarce (114, 221, 238, 239), aside from the suggestive structural data (153, 282), which are reviewed in reference 170.
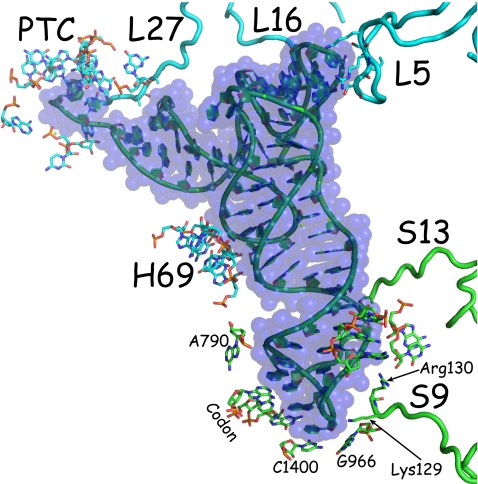
The peptidyl-tRNA, unlike the aminoacyl-tRNA in the A site, makes many interactions with various moieties in the P site, the majority of which are to 16S and 23S rRNA and only a few of which are to ribosomal proteins, which will be considered first (Fig. 2). Amino acids of three large-subunit proteins (L5, L16, and L27) and two small-subunit proteins (S9 and S13) make direct contacts with the peptidyl-tRNA (170). The C-terminal ends of two proteins, S9 and S13, extend toward the anticodon loop of the peptidyl-tRNA (57, 347, 358). For S9, the C-terminal Arg, which is conserved among Bacteria, contacts the 5′-phosphate of nucleotide 32 of the peptidyl-tRNA, and the length of this protein is invariant. The second-to-last amino acid is a conserved Lys, which contacts the 5′-phosphates of nucleotides 33 and 34 (282). The C terminus of S13 phylogenetically varies in length, but it always contains several basic side chains. These interact with the backbone of peptidyl-tRNA, since it runs parallel to the anticodon stem. Various deletions of the C-terminal ends of S9 or S13 reduce peptidyl-tRNA binding (136), suggesting that these proteins may be engaged to maintain the grip of peptidyl-tRNA in the P site and thereby may also contribute to standard reading frame maintenance. Indeed, deficiency of, or alterations in, the C termini of ribosomal protein S9 induce +1 frameshifts, consistent with a role of this protein in maintaining the reading frame (221). The large ribosomal protein L5 interacts with the D arm and TΨC loop. L27 and L16 interact with the acyl stem of peptidyl-tRNA. From an evolutionary perspective, precursors to these proteins may have entered more RNA-based ribosomes to facilitate massive speed, enhancing utilization of EF-G-mediated translocation (69, 99, 108, 109, 252, 290). Though many studies have pointed to translocation being an intrinsic feature of ribosomes (see, e.g., reference 94), several have shown that it is the RNA components of the ribosome that are critical for this reaction (225, 226).
FIG. 2.
tRNAfMet in the P site of a bacterial 70S ribosome (282). Protein and rRNA residues of the 30S (with C atoms in light blue) and 50S (with C atoms in darker blue) ribosomes that have atoms within 3.8 Å of the peptidyl-tRNA are shown as stick representations and the protein chains as tubes (blue from L50 and green from S30). The image was made created by use of PyMOL (82). The last (Arg130) and the next-to-last (Lys129) amino acids are indicated. (Courtesy of J. Näsvall, Umeå University, Umeå, Sweden.)
In the 30S A site, only four nucleotides of 16S rRNA contact tRNA, and three of them (A1492, A1493, and G530) make contacts with the anticodon nucleotides (241, 282), whereas 16S rRNA makes 10 interactions with P-site tRNA and none directly with the anticodon bases (32, 282). The 16S rRNA-tRNA interactions, observed in the three-dimensional structure, are in agreement with the earlier established protection and modifications studies (reviewed in references 113 and 170). The anticodon stem contacts the backbone of three nucleotides (1229, 1230, and 1341) and by two bases (G1338 and A1339). However, the conformation of the anticodon is stabilized by stacking interactions between m5C1400 (C1400) and the wobble base and packing of m22G966 toward the ribose of the wobble nucleoside (33, 256, 282, 300). Thus, there is an extensive stabilization of the space orientation of the wobble nucleoside by 16S rRNA moieties but not of the other two nucleotides of the peptidyl-tRNA anticodon. This is in marked contrast to the Ramakrishnan A-minor rRNA calibration, by minor groove sensing, of the anticodon pairing of the first two codon bases in the ribosomal A site (241, 282). The absence of such calibration is likely critical for P-site codon-anticodon dissociation, which is central to at least most models of ribosomal frameshifting (24, 107, 187, 259, 304, 320). Dissociation of P-site codon-anticodon pairing is not, however, sufficient for frameshifting. Though contacts by the backbone of the P-site codon with three conserved 16S rRNA nucleotides (G926, m3U1498, and m4Cm1402) may help to fix the mRNA position for standard decoding, it can evidently be overridden, permitting the selective advantage of programmed frameshifting.
Whereas aminoacyl-tRNA makes only a few direct interactions with 23S rRNA in the A site (an H bond between the ribose of A1913 and ribose 37 [282] and a base pair between C75 and the A loop of the ribosome [50, 161, 224]), there are numerous nucleosides in domains IV and V of 23 S rRNA that are protected by peptidyl-tRNA from chemical probing, suggesting that it is close to these domains of 23 S rRNA. Specifically, the CCA terminus makes direct contacts with A2451, G2251, and G2252 of 23S rRNA, suggesting that its position is strongly influenced by 23S rRNA (31, 270, 282, 285).
The ribosome induces pronounced structural changes of the tRNA when it traverses between the A, P, and E sites. In the P site, the body of the P-site tRNA has a kink at the junction of the anticodon- and acceptor-stem domains, resulting in a 10°C bend of the tRNA body relative to the ASL. Moreover, the anticodon of the P-site tRNA is also changed compared to that of tRNA in solution (282, 300) (see Fig. 5b of reference 170). Therefore, the anticodon-codon interaction in the P site is quite different from that occurring in the A site.
The ribosomal structure features described above set the scene for the framing issue. Peptidyl-tRNA interacts with both 16S and 23S rRNAs and several ribosomal proteins at numerous sites, suggesting that the position of the peptidyl-tRNA is more dependent on such interactions than on interactions with the anticodon. Clearly, the ribosomal “grip” of peptidyl-tRNA is determined by features outside the anticodon, in contrast to the binding of aminoacyl-tRNA in the A site, which is almost solely dependent on interactions of the anticodon with mRNA and rRNA. These numerous interactions between peptidyl-tRNA and the ribosome suggest that they may be an important parameter to maintain the reading frame. If so, we would expect that changes in at least some of these interactions, caused by alteration of the tRNA or in the ribosome, may induce frameshifting. Distortions may lead to tRNA losing its grip on mRNA, which could then slip. However, an alternative is via altered translocation step size.
MORE EFFICIENT +1 FRAMESHIFTING WHEN THERE IS POTENTIAL FOR FOUR-BASE ANTICODON-CODON PAIRING: ALTERNATIVES TO A QUADRUPLET TRANSLOCATION MODEL
As predicted by the original ribosomal A- and P-site model (329), peptidyl-tRNA translocates from the A to the P site, and this movement is catalyzed by EF-G (90, 329). Various models have been presented to account for how this movement is achieved (80, 123, 148, 209, 226, 267, 279, 292, 293, 348, 359). Although they differ in detail, they feature the tRNA-mRNA complex moving from the A to the P site as a unit. This view was strengthened by experiments showing that peptidyl-tRNA (AcPhe-tRNA) cross-linked to mRNA in the A site can be transported as a unit from the A to the P site (202). Since the anticodon consists of three nucleotides (140), one model involved the length of mRNA movement being determined by the number of anticodon bases involved in pairing with it. On the basis of this model, if the size of the anticodon could be increased, codon size might correspondingly increase. We will next consider whether tRNA mutants which cause +1 frameshifting, many of which have expanded anticodon loops (summarized in Table 1), have four bases which function as an enlarged anticodon in the A site.
TABLE 1.
+1 frameshifts induced by altered tRNAs
| tRNA or stress | Alteration in tRNA | Site of frameshifta | Expt type | Reference(s) |
|---|---|---|---|---|
| S. enterica tRNAGGGPro | Base substitutions outside and in anticodon; tRNA aminoacylated as in WT | P site, by WT tRNAcmo5UGGPro | In vivo | 258, 259, 295 |
| S. enterica tRNACGGPro | Insertion of an extra nucleotide in anticodon | P site, by either mutated tRNACGGPro or WT tRNAcmo5UGGPro | In vivo | 259 |
| Yeast tRNAIGGPro | Insertion of an extra nucleotide in anticodon | P site | In vivo | 259 |
| Yeast tRNACCCGly | Insertion of an extra nucleotide in anticodon | P site | In vivo | 259 |
| S. enterica tRNAmnm5UCUArg | Alterations in structure of tRNAmnm5UCUArg reduce concn of aminoacylated tRNAmnm5UCUArg, which in turn causes frameshift at next upstream codon | P site | In vivo | 180 |
| Starvation | Amino acid limitation | P site | In vivo | 107 |
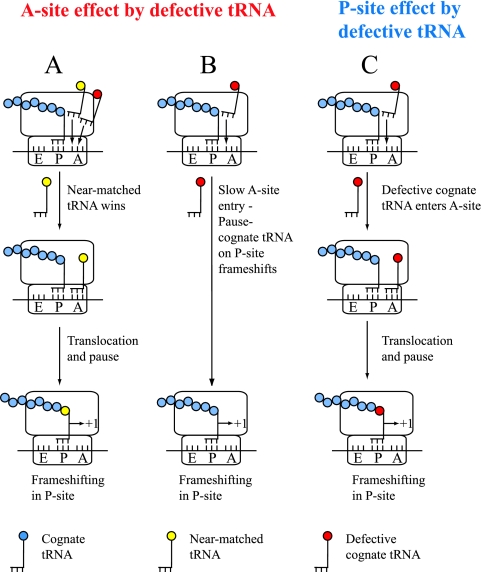
| Third-position-mismatched tRNAs | Lack of cognate tRNAs induces +1 frameshift by near-matched tRNA; i.e., imbalance of tRNAs causes +1 frameshift | P site | In vivo | 304 |
| Imbalance of tRNAcmo5UGGPro or tRNAmnm5UCUArg | Overexpression of tRNAcmo5UGGPro induces +1 frameshift; +1 frameshift at tandem AGG-AGG caused by limitation of tRNAmnm5UCUArg | P site | In vivo | 229, 291 |
| Third-position-mismatched tRNAs | Several third-position-mismatched tRNAs in P site induce frameshift | P site | In vivo | 304 |
| tRNA modifications | Various deficiencies in tRNA modification cause +1 or −1 frameshifts in E. coli, S. enterica, or yeast | In some cases shown to occur in P site; in other cases not addressed | In vivo | 40, 120, 176, 188, 320, 325 |
| E. coli tRNAmnm5UCCGly | Base substitutions in position 34 and outside anticodon | P site | In vivo | 131 |
| E. coli tRNAcmo5UACVal | Insertion of either A or U in anticodon between cmo5U34 and A35 | P site | In vivo | 235 |
| S. enterica tRNAGGUThr | Insertion of an extra nucleotide in 5′ part of the anticodon and “pushing” U33 into wobble position, resulting in 5′-UUGG-3′ anticodon | Possible P site (see text) | In vivo | 45, 46 |
| S. enterica tRNAcmnm5s2UUGGln | Overexpression of tRNAcmnm5s2UUGGln with an inserted U between cmnm5s2U34 and U35 | Not experimentally addressed | In vivo | 228 |
| S. enterica tRNAmnm5s2UUULys | Overexpression of tRNAmnm5s2UUULys with an inserted U between mnm5s2U34 and U35 | Not experimentally addressed | In vivo | 228 |
| E. coli tRNAICGArg | Changes in or outside anticodon | Not experimentally addressed | In vivo | 317 |
| E. coli tRNACUALeu | Insertions of an extra nucleotide in anticodon and A26G base substitution | Not experimentally addressed | In vivo | 211 |
| E. coli tRNAcmo5UACVal | Insertion of an extra nucleotide (C32.1) and G42A present on plasmid | Not experimentally addressed | In vivo | 230 |
| E. coli tRNAcmo5UACVal | Base substitutions (C74A or C74G) in the CCA terminus | Not experimentally addressed | In vivo | 239 |
| Mitochondrial tRNAUGASer | U42C and lack of Ψ27. | Not experimentally addressed | In vivo | 143 |
| tRNAAla | Insertion of an extra nucleotide in anticodon; in vitro experiments | Not experimentally addressed | In vitro | 326 |
| tRNAPhe | Insertion of CCCG as anticodon in body of unmodified tRNAPhe; in vitro experiments | Not experimentally addressed | In vitro | 253 |
The site of frameshifting was determined as described in the text under Tests of P-Site and E-Site Roles: Genetic Evidence for E-Site Codon-Anticodon Pairing.
Derivatives of Glycine tRNAs: the Finding of More Efficient +1 Frameshifting when Four Codon Bases Are Complementary to Four tRNA Bases
E. coli sufD42 is a derivative of tRNACCCGly with an extra C in its anticodon loop (262). (It has no modification in its anticodon loop, unlike the other GGG-reading tRNA, tRNAmnm5UCCGly, which has mnm5U34 as its wobble nucleoside. It is also known as tRNA1Gly and is encoded by glyU.) The simplest model to explain how it suppresses +1 frameshift mutants with runs of G is that four rather than three bases constitute the anticodon. Pairing of these four bases with four codon bases in the A site leads to quadruplet translocation (134, 262), thereby restoring some ribosomes to the WT frame. This was the main justification for the “yardstick” model (see above). If such an interaction involved simultaneous Watson-Crick pairing by four mRNA bases to tRNA, then there are simple predictions for the specificity of tRNA suppressors of +1 frameshift mutants. This was explored in S. cerevisiae.
The yeast SUF16-1 suppressor is a derivative of tRNAGCCGly with an extra C in its anticodon loop. The anticodon has G34 as a wobble nucleoside and the extra C is at position 36.5, resulting in the sequence GCCC. Making specific alterations in the first position (NCCC, where N is any of the four nucleosides) revealed that suppression does not require a base pair between the N34 and the cognate nucleoside in the mRNA, although its efficiency is enhanced by the potential to form Watson-Crick pairing (104). The conclusion about the potential for four mRNA bases being able to form such pairing, with tRNA enhancing but not being required for this type of +1 frameshifting, was also reached with E. coli sufD (334). However, the yeast work was the first to cleanly show it.
The ribosomal site at which suppressor tRNA mediates frameshifting is apparent from work with other S. cerevisiae suppressors and other tRNA mutants described below. The S. cerevisiae suppressors SUF3 and SUF5 are derivatives of another Gly-tRNA (tRNACCCGly (77). Overexpression of the tRNA reading the codon downstream of the GGGG quadruplet reduces the frequency of frameshifting, and if a stop codon is downstream of the frameshift site, the efficiency of frameshifting increases. These results demonstrate that the frameshift event occurs in the P site (259). Though the ribosomal site at which E. coli sufD42 mediates frameshifting has not been addressed, one relevant experiment has been performed. sufD42 was tested for its ability to suppress a −1 frameshift mutant, with a positive result (334). While this could reflect different mechanisms, it is tempting to deduce that it reveals different aspects of just one P-site based mechanism.
Derivatives of Proline tRNAs: a Focus on Events in the Ribosomal P Site
The sufA6 and sufB2 frameshift suppressors of S. enterica are derivatives of tRNACGGPro and tRNAGGGPro with an extra G in their anticodon loops. In the WT these tRNAs have m1G37 next to and 3′ of the anticodon, and both of these frameshift suppressors have this modification at a position which places the extra G such that the anticodon consists of three nucleotides bordered by U33 and m1G37 (259). Since m1G37 prevents Watson-Crick base pairing (222), these frameshift suppressors are unable to make quadruplet base pairing. Moreover, the frameshift occurs in the P site, since the efficiency of both sufA6- and sufB2-induced frameshifting is sensitive to the efficiency of decoding the next downstream codon (258, 259). The tRNA causing the frameshift event in the sufB2 mutant is the third-position-mismatched tRNAcmo5UGGPro (259). Still, in some contexts, the sufB2 tRNAGGGPro itself promotes frameshifting, suggesting that the altered structure of the sufB2 tRNAGGGPro is also prone to reposition in the P site (259). With sufA6-induced frameshifting, up to 50% of it is caused by the third-position-mismatched tRNAcmo5UGGPro, but the rest of the frameshift event is apparently caused by the altered sufA6 tRNACGGPro (259). This suppressor is a derivative of proK tRNACGGPro with an extra G in its anticodon loop, and it suppresses at the site C-CCC-UGA (hisD3749). It is not clear how this tRNA can out-compete the proL tRNAGGGPro, whose cognate codon is CCC and which is also present in the cell (259). Nevertheless, sufA6-mediated suppression occurs in the P site (259). The sufA and sufB results provided strong evidence against the yardstick model (259).
In Salmonella, deficiency of m1G37, which is present in all three proline tRNAs, results in suppression of +1 frameshift mutations in a run of Cs (40). Though the anticodon loop does not have an extra base, the absence of m1G37 creates the potential for a four-base anticodon with unmodified G37 making a Watson-Crick base pair with an extra C at the frameshifting site. Initially it was postulated that a four-base interaction occurs in the A site, with a four-base translocation positioning the ribosome in the correct reading frame and explaining how the lack of m1G37 suppresses certain +1 frameshift mutations (40, 120). However, the frameshift event occurs in the P site (258), making it unlikely that quadruplet base pairing in the A site mediated by m1G37 deficiency is a prerequisite for the frameshift event.
In S. cerevisiae both the SUF2 and the SUF10 genes encode tRNAIGGPro. Their suppressor derivatives (SUF2-1 and SUF10-1) have an extra G in the anticodon loop (76). The WT form of these tRNAs has m1G37 next to and 3′of the anticodon, but whether the suppressor derivatives retain the m1G37 is not known. This issue is relevant, since the presence of the methyl group of m1G37 would prevent Watson-Crick base pairing required for a quadruplet base pairing by these mutated tRNAs. However, SUF10-1 causes frameshifting in the P site (259).
Taken together, these results suggest that the frameshift event occurs when the classical frameshift suppressors derived from proline tRNAs in Salmonella and in S. cerevisiae are in the P site rather than the A site.
Derivatives of Threonine tRNA: a +1 Suppressor That Does Not Act at a Run of Repeated Bases
The S. enterica sufJ frameshift suppressor is a derivative of tRNAGGUThr (also denoted tRNA3Thr) whose sequence and modification pattern have not been determined. The sufJ128 derivative has an extra C in the 5′ side of the anticodon, “pushing” the U33 nucleoside into the wobble position 34 and theoretically changing the anticodon from 5′-GGU-3′ to 5′UGGU-3′. This suppressor reads the quadruplet ACCX sequence (where X can be A, U, or C) (45). It suppress the hisG6608 mutation (the sequence around the frameshift mutation is ACCCUGC [44]; ACC is a Thr codon, and CUG is a Leu codon). Its ability to suppress hisG6608 is dependent on the presence of a truA (hisT) mutation, whose gene product is responsible for the synthesis of Ψ in the anticodon stem and loop in a subset of tRNAs. Since tRNAGGUThr is not a substrate for the TruA enzyme, the truA-mediated increase in the activity of sufJ must be caused by another tRNA which normally has Ψ, very likely in its anticodon loop or stem. In the hisG6608 context, tRNACAGLeu, which has Ψ in positions 38 (loop) and 40 (stem), decodes the zero-frame codon, CUG, which follows ACC. This CUG overlaps the 3′ base of the ACCC at which sufJ tRNA causes frameshifting. Lack of Ψ in tRNACAGLeu reduces its ability to enter the CUG primed ribosomal A site (185). sufJ tRNA presumably competes with the tRNACAGLeu for the underlined C in the sequence ACCCUGC. It has been proposed that when the less efficient Ψ-deficient tRNACAGLeu is present, sufJ tRNA has a higher probability of interacting with the four nucleotides ACCC in the A site, thereby allowing the next tRNA to read the +1 frame UGC codon (44). However, these data are also consistent with a normal three-nucleotide translocation by sufJ tRNA and its mediation of frame change occurring in the P site. In this scenario decreased efficiency of decoding of the A-site CUG by incoming aminoacyl tRNA, due to Ψ deficiency of tRNACAGLeu, will increase the probability of sufJ tRNA changing frame in the P site. The available genetic data do not allow a distinction between this model and the four-base A-site codon model.
Derivatives of Glutamine tRNA: U-Rich Anticodon and Specificity of Suppression Site
The sufG suppressors in S. enterica and E. coli are derivatives of tRNAcmnm5s2UUGGln (228). The sufG tRNA has an extra U in its anticodon, thus expanding the anticodon loop to eight nucleotides and having an apparent anticodon sequence of cmnm5s2U34-U35.5-U35-G36 (228). These derivatives of tRNAcmnm5s2UUGGln apparently suppress at the sequence CAAA but not CAAU, CAAC, or CAAG (228). Since the cmnm5s2U34 is suggested to be restricted in its ability to wobble toward G, the inability of the suppressor to decode CAAG is reasonable. However, this restrictiveness induced by the modification has been questioned (37, 119), and in fact, a similar derivative (mcm5s2U) of this kind of modification promotes reading of G-ending codons (154). Moreover, since the suppressor tRNA was overexpressed, the inability to decode CAAG was unexpected. Peptide analysis of the frameshift product revealed that only one amino acid, Gln, was incorporated at the CAAA site, consistent with four anticodon bases simultaneously pairing with four codon bases and with the mutated tRNA causing the frameshift event (228). However, the data do not permit distinction between the frameshift occurring by quadruplet translocation from the A site to the P site or the aberrant tRNA positioning itself into the +1 frame when present in the P site after a normal three-nucleotide translocation. Repositioning in the P site by the altered tRNA may be sequence sensitive. An anticodon loop with as many Us as are present in this tRNA derivative is most likely very flexible due to the poor stacking potential of U. Therefore, an unusual conformation of the anticodon loop cannot be ruled out and could explain the inability to suppress the CAAG/U/C sites.
Derivatives of Lysine tRNA: U-Rich Anticodon Stacking Issues Similar to Those for sufG
A frameshift suppressor derivative of tRNAmnm5s2UUULys enhances frameshifting at the sequence CAA AAA ACC (228). The altered tRNA has an insertion of G between U36 and t6A37, expanding the anticodon loop to eight nucleotides and creating an apparent anticodon sequence of mnm5s2U34-U35-U36-G36.5 (the modification status was not established). As for the sufG suppressors described above, this lysine-specific suppressor suppresses only frameshift mutations having the sequence CAAA close to the frameshift mutation, and only Lys was incorporated at the position corresponding to the CAAA sequence. The inability to decode CAAG is surprising. No other lysine tRNA exists, and an overexpressed suppressor tRNA having the anticodon mnm5s2U34-U35-U36-G36.5 should also be able to make a four-anticodon base pairing with CAAG. Note that this tRNA derivative has an unusual U-rich anticodon, making it likely that when undermodified it is even more flexible than pointed out for the tRNAcmnm5s2UUGGln (see above). A critical issue in this respect, as well as for sufG-mediated suppression, is whether four anticodon bases simultaneously pair with four codon bases and if so at which ribosomal site(s). The answer is unknown, but modification at the first anticodon nucleotide is likely relevant.
Derivatives of Valine tRNA: Base Insertion in the Anticodon Leads to +2, +3, or +6 Decoding
The mutants of GUG-, GUU-, and GUA-decoding E. coli tRNAVal with an extra base, A or U, at position 34.5 within the anticodon are briefly included here because of the similarity of this feature to those of mutants in the other categories. However, these mutants, termed hopR, were isolated for their ability to suppress the −1 frameshift mutation trpE91 and do so by causing +2 frameshifting at the sequence GUGUGA (235). Since only one amino acid, valine, was encoded by the first five nucleotides, dissociation of anticodon pairing with the underlined GUG and re-pairing to mRNA at the GUG in italics were inferred. With the sequence GUU UAA GUU, these mutants cause hopping onto or over the stop in an anticodon-dependent manner. This aspect will be considered below because of its relevance to alternative stacking considerations. Here the relevant feature is that the mutant tRNAcmo5UACVal has to be in the P site as it mediates these events, since the stop codon is sensed in the A site.
Derivatives of tRNAICGArg Causing Frameshifting at a Proline Codon: Seven-, Eight-, or Nine-Nucleotide A/C Loops
An external suppressor (sufT621) (15) of the +1 frameshift mutant trpE872 is a derivative of tRNAICGArg (also denoted tRNA2Arg) containing an extra G between position 36 and 37 (317). This sufT621 tRNA has an eight-nucleotide anticodon loop with the anticodon U33-ACGG-A37. Modifications at positions 34 and 37 were not determined, but its WT form contains inosine (I) in position 34 and m2A37. Protein sequence analysis established that the frameshift event occurs at the border of CGU (Arg) and AUU (Asn) present in the sequence AAC-CCG-U-AUU. The potential four-nucleotide anticodon ACGG is thus complementary to the CCG-U sequence at which the frameshift occurs. Changing the first C of CCG-U to any other nucleoside abolishes suppression, whereas changes of U did not influence frameshifting, demonstrating that suppression does not require perfect complementarity at the “wobble position.” However, it is sensitive to the nature of the interaction involving the first base of this four-base codon. Since a quadruplet interaction apparently is not a requirement for suppression, it is not necessary to postulate four-nucleotide base pairing in the A site. One possibility is that the tRNAICGArg reads the proline codon CCG, and after translocation, the altered tRNAICGArg shifts frame from CCG to CGU. If so, overexpression of tRNACGGPro (CCG decoding) or tRNAQUATyr (UAU decoding) should inhibit suppression. Other mutants of the same tRNA predicted to have nine or even seven nucleotides in their anticodon loops also caused the +1 frameshifting (317).
Analysis of In Vitro-Constructed tRNAs or ASL with an Expanded Anticodon Loop
Incorporation of nonnatural amino acids is sometimes desired to create new proteins and to facilitate structural studies (327). One way to achieve such altered proteins would be to construct tRNAs able to accept nonnatural amino acids and to read a four-base codon. Four-base codons have successfully been used to incorporate nonnatural and/or normal amino acids into proteins at desired sites (7, 138, 194, 210, 211). In general, maximum efficiency of incorporation of such nonnatural amino acids was achieved when there was potential for four anticodon bases to pair with four codon bases. The analysis did not distinguish whether the complementarity was required at the ribosomal A or P site. However, recently, this question has been addressed in vitro (253, 326) using either synthesized tRNA or ASL (anticodon-stem-loop) constructs with an eight-nucleotide anticodon loop. The movement of the tRNA on the ribosome was analyzed by toe printing, which revealed net translocation by four nucleotides from the A site to the P site. Since such analysis shows the final position of the tRNA but does not show how this occurs, two possible interpretations can be made. The 3′ three nucleotides of the flexible anticodon pair in the A site with the 5′ three nucleotides of the four nucleotides constituting the frameshift site on the mRNA. A four-base-pair helix between the expanded anticodon and the four-nucleotide frameshifting site is formed during translocation, thus inducing a quadruplet translocation (326). Alternatively, a three-base-pair interaction occurs in the A site, and following a normal three-nucleotide translocation, repositioning occurs when the tRNA-mRNA package lands in the P site, thereby moving the P/A boundary +1 nucleotide (211, 259). A toe print assay would give the same result, but the mechanism would be different.
An Expanded Anticodon Loop Can Be Accommodated into the A Site without a Fourth Potential Anticodon Base Being Involved in Codon Pairing
Three different ASLs with expanded anticodon loops were soaked into the A sites of crystallized ribosomes containing mRNA with four complementary bases (88). In all cases the ASLs were accommodated into the A site, but interestingly, in no case were four bases observed to be involved in Watson-Crick base pairing (Fig. 1A and B). Nevertheless, novel noncanonical interactions were discovered, consistent with a four-base occlusion model (46). However, as discussed above, it is unclear whether these structures in the A site per se are relevant for frameshifting, since the toe print assay for translocation monitors the final position in the P site and not necessarily a quadruplet translocation.
In summary, several data related to frameshift suppression induced by tRNA mutants with an extra nucleotide in the anticodon loop are inconsistent with a requirement for quadruplet base pairing in the A site. Derivatives of Gly- and Pro-tRNAs with an eight-nucleotide anticodon loop, in both bacteria and in yeast, shift frame in the P site, making it unnecessary to postulate four-base pairing in the A site as suggested by the yardstick model. For other frameshift suppressors the data do not rule out quadruplet base pairing in the A site, but their action can be explained without invoking it.
DOES FRAMESHIFTING BY −1 FRAMESHIFT MUTANT SUPPRESSORS DEPEND ON DOUBLET PAIRING IN THE A SITE?
In at least the great majority of cases, anticodon-codon pairing, its dissociation, and anticodon re-pairing to mRNA at an overlapping codon in the −1 frame is central to −1 frameshift mutant suppressors mediating their effect. Members of one bacterial suppressor class, sufS, are mutants of tRNAmnm5UCCGly (tRNA2Gly) (246), whose WT form is the only decoder of GGA in vivo; it also reads GGG (216). The WT anticodon sequence is mnm5UCC (T. Suzuki [personal communication] determined the identity of the modification). In S. enterica or E. coli where the tRNA is identical, just replacement of mnm5U by C (the sup-601/sufS601 suppressor (246), causes significant −1 frameshifting at the sequence G GGA (where GGA is in the zero frame). It is this mutant tRNA with the CCC anticodon, and not some related tRNA, that performs the frameshifting (131, 249). Though the mutant strain has no tRNA matched to the GGA codon, it grows well. The anticodon of WT tRNA1Gly is also CCC, but it differs at several positions elsewhere in the tRNA. Perhaps a crucial difference for adequate triplet reading of GGA necessary for detectable cellular growth, however, is that WT tRNACCCGly (tRNA1Gly) has U at position 32, whereas tRNAmnm5UCCGly (tRNA2Gly), and thus the mutated form of it, has C at position 32. Since C32 is not discriminatory (61, 192, 244), the sufS mutant form of tRNAmnm5UCCGly with its anticodon CCC must read GGA sufficiently well for viability. (The other differences between this mutant and WT tRNACCCGly [tRNA1Gly] may be irrelevant to the frameshifting, but this has not been tested.) The anticodon pairing properties of tRNAmnm5UCCGly are such that its base 34 can be replaced with C so it reads a third-position-mismatched codon in addition to its original cognate codon, GGA, to directly mediate the most efficient −1 frameshifting of all the isolated mutants of this tRNA (the paragraph after the next deals with other mutants).
The nature of the sequence at which the mnm5U-to-C wobble base-substituted tRNAmnm5UCCGly (tRNA2Gly) mutant causes frameshifting has been investigated. Replacing ACC 3′ adjacent to GGA with the rare codon AGG or a stop codon increases the level of frameshifting, suggesting that at least the majority of the frameshifting occurs in the P site. Replacing the 5′ G of the G GGA sequence with A or C also causes a large reduction in frameshifting, consistent with re-pairing to mRNA in the −1 frame rather than doublet pairing in the A site being the key event (131). (In these experiments a five- to sevenfold decrease was detected, which is greater than those in an earlier experiment [245] and in an experiment when its encoding gene was expressed from pACYC184 [227].) Further evidence for a detachment and slippage mechanism comes mainly from the direct ability of the mutant tRNAs in E. coli to mediate stop hopping, i.e., detaching from the initial GGA in the sequence GGA UAA GGA and, following mRNA slippage re-pairing to mRNA at the second GGA (or preferably GGG), allowing coding resumption at the following codon after specification of a single amino acid, glycine, by the nine nucleotides (131). The simple conclusion is that at least most of the time, the key event for frameshifting occurs in the ribosomal P site following triplet translocation. However, the nature of the decoding of the third codon base, the A of GGA, in the A site is unclear. (At an early stage, the potential for alternative structures of the D arm of the related phage T4 glycine tRNA [26] and with the E. coli tRNA for tautomerization of C34 to the imino form, which could allow C·A pairing [311], was pointed out.) A case of programmed translational bypassing of 50 nucleotides (344) involves the same glycine tRNA being the peptidyl-tRNA during the bypassing. All mutants genetically selected to have decreased bypassing efficiency had alterations in this glycine tRNA and outside of its anticodon loop (129). In a limited study, most but not all of these mutants, termed byp, plus sufS suppressors other than the sufS601 suppressor just considered, showed that WT tRNACCCGly (tRNA1Gly) can also mediate frameshifting at GGA (131). In these sufS mutants of tRNAmnm5UCCGly (tRNA2Gly), the alterations are outside the anticodon arm, and the amount of the aminoacylated product tRNA is substantially but differentially reduced (131, 246). The efficiency of these suppressors is lower. Since tRNACCCGly (tRNA1Gly) has U at position 32 and so is discriminatory, the nature of the A-site decoding may be different from that just discussed. Very relevant to this class of suppressors are earlier studies of a multicopy clone of WT DNA which acts as yet another suppressor of the −1 frameshift mutant trpE91. Its active segment encodes WT tRNACCCGly (tRNA1Gly) (227) (see below). Instead of WT tRNACCCGly competing with debilitated tRNAmnm5UCCGly, excess tRNACCCGly competes with unperturbed levels of WT tRNAmnm5UCCGly to mediate −1 frameshifting at GGA (227). An increased efficiency of frameshifting with a G, rather than U, C, or A, 5′ of the GGA at which the −1 frameshifting occurs is not apparent except when U32 in the WT tRNACCCGly (tRNA1Gly) is replaced by C (227) (but note that in this series of experiments, the enhanced effect of a 5′ adjacent G seen with sufS601 mutant tRNAmnm5UCCGly [tRNA2Gly] is less than that reported in reference 131). Mycoplasma glycine tRNA has C32, which when replaced by U discriminates among glycine codons according to wobble rules, whereas the WT tRNA reads all four glycine codons (61, 192). This effect is known only for glycine tRNAs, though other tRNAs have C32. Whether the explanation hinges on kinetics, some unusual pairing, or lack of it, as in the two-out-of-three mechanism proposed for Mycoplasma tRNA by Lagerkvist (173), is unknown. If the −1 frameshifting mediated by tRNACCCGly (tRNA1Gly) at GGA predominately follows triplet translocation, as that mediated by sufS601 mutant tRNAmnm5UCCGly (tRNA2Gly) appears to, then there is the issue why the level of the frameshifting that it mediates is not higher when there is the potential in the P site for its anticodon dissociating from mRNA and re-pairing to it at the −1 frame GGG. One possible explanation is that only the first two codon bases pair with tRNA in the A site and doublet translocation ensues. Another possible explanation is that lack of pairing of the third codon base by P-site tRNA allows that third mRNA base to instead pair with the incoming A-site tRNA. Whether this would involve the first two P-site codon bases remaining paired to their original counterpart tRNA bases or being dissociated from the anticodon of P-site tRNA is unspecified. Protein synthesis can proceed without P-site tRNA anticodon being paired to mRNA. Apart from much early in vitro work, −1 frameshifting has been shown in vivo in special circumstances to occur without re-pairing (188) and is also known from bypassing studies (132). However, its efficiency is low, and the alternative possibility of doublet decoding merits attention.
The other altered tRNA in which mutations were isolated on the basis of their ability to cause −1 frameshifting is GUG/GUA and GUU-decoding tRNAVal. Two types of mutants were isolated, with one class being substitutions of C74, the first C of the universal CCA at the 3′ end of tRNAs (239). As considered below, members of the other class have an extra base in their anticodon loops. Protein sequence analysis of the products of their action at several suppressible sites has been done (235). The studied frameshifting that they cause involves P-site anticodon dissociation from, and re-pairing to, mRNA. Though their anticodon pairing in the A site may be exceptional, the key relevant event is in the P site.
Also, as shown by one of us (334), sufD, in addition to classical +1 frameshift mutant suppression, can also suppress a −1 frameshift mutant. This is likely another aspect of its proposed P-site frameshifting.
In summary, in most of the cases described, the key events for at least the substantial majority of frameshifting are concluded to be in the P site following triplet translocation. However, the simplest interpretation of one case involves doublet translocation, but this is not proven.
TESTS OF P-SITE AND E-SITE ROLES: GENETIC EVIDENCE FOR E-SITE CODON-ANTICODON PAIRING
As presented above, many of the suppressor tRNA-mediated frameshift events occur after the A site (for reviews of non-suppressor-mediated post-A-site frameshifting, see references 24, 93, 187, 219, and 297). However, the suppressor data on its own are inadequate to ascertain whether in some cases A-site events followed by translocation of a nontriplet codon are involved. Nevertheless since post-A-site events almost inevitably involve codon-anticodon realignment, or at a minimum pairing/occlusion of one additional base or one fewer base, the nature of A-site decoding is highly likely to influence the efficiency, as has also been demonstrated with several cases of programmed frameshifting. For +1 and +2 frameshifting, slow decoding of the A site delays sequestration of the 5′ mRNA base(s). It also provides greater time for either −1 or +1/+2 frameshifting. Two kinds of experiments have been performed to address efficiency of zero-frame A-site decoding on suppressor-mediated frameshifting. Where a sense codon is involved, this rate can be either increased (usually by overexpression of cognate tRNA reading the A-site zero-frame codon) or reduced (by reducing the number of genes encoding the tRNA or by altering its ability to be accepted at the A site, e.g., by modification deficiency [185]). For six alterations of different tRNAs, the frameshifting has been shown to occur in the P site by such experiments (Table 1). (Since stop codons are slow to decode, in another case, the codon 3′ of the shift codon was constructed to be a stop codon [131].) Note that in several cases the mutated tRNA is a classical frameshift suppressor with an expanded anticodon loop. Thus, frameshifting does not occur by quadruplet translocation even if four mRNA bases may be involved in tRNA pairing in the A site (derivative of yeast tRNAIGGPro and tRNACCCGly [259]).
Irrespective of whether WT or mutant translation components are involved in slippage-mediated frameshifting, the term “peptidyl-tRNA slippage model” has been quite widely used for the relative anticodon-mRNA movement involved (28, 122, 259, 320). Alternatively, a third-position-mismatched peptidyl-tRNA (or an otherwise nonoptimal peptidyl-tRNA such as mutated or modification-deficient tRNA) distorts the A site in such a way that out-of-frame binding of aminoacyl-tRNA occurs in the A site and peptidyl-tRNA does not detach from the mRNA (93, 96, 123, 298, 304). This is known as the “out-of-frame binding model” or “once-only-pairing” model and continues to be the source of a debate about different ways that it could happen and its validity. Note, however, that in both models the frameshift is induced by the peptidyl-tRNA, either by its slippage relative to mRNA or by its induced distortion of the A site mediated by suboptimal binding.
Structural studies have revealed altered tRNA conformations during translation compared to the structure free in solution. Any mutational assessment of the functional importance of the different conformational changes, including in relation to framing, is not a simple challenge given tRNA's compact nature and multiple interactions. Further, debilitation of tRNA, including alterations to aminoacylation and modification efficiency, may facilitate decoding and frameshifting by WT tRNA with the third position mismatched rather than by the mutant tRNA itself. Determination of whether tRNAs with their alterations outside of the anticodon loop directly mediate frameshifting or whether the debilitation facilitates decoding and frameshifting by a third-position-mismatched tRNA, or a combination of the two, has been performed in several cases. Debilitation of a tRNA can lead to the decoding of its cognate codon being performed part of the time by a related tRNA which also mediates one type of codon realignment, but when the debilitated tRNA itself does the decoding, it can be prone to a different type of realignment. Likely examples of this are several mutants of GGA-decoding tRNAmnm5UCCGly (tRNA2Gly) outside of its anticodon. The −1 frameshifting in strains with these mutants is largely caused by tRNACCCGly (tRNA1Gly), whereas the forward hopping can best be explained as predominately involving the mutant tRNAmnm5UCCGly (tRNA2Gly) (131). Details about the nature of the debilitations are given below, but while inferences in relation to structure can be made, determination of relevance for only partially understood conformational changes during translation is not attempted. This reticence does not, however, apply to substitution mutants of C74, the first C of the CCA at the 3′ end of tRNA to which the amino acid is attached. The mutants, which are of E. coli tRNAcmo5UACVal, were isolated by M. O'Connor as suppressors of the −1 frameshift mutation trpE91 (239). The compensatory frameshifting that they cause is probably +2 (239). The suppressors have their single mutation in one of the four identical genes for this tRNA. Accordingly, the mutant tRNA is able to effectively compete with WT tRNA, even though the 5′ base of the universally conserved CCA at the 3′ end of tRNA in different suppressors is replaced by either A or G. (This finding that C74 could be replaced without drastic consequences occurred just before the major discovery that C74 and C75 of the CCA terminus base pair with G2252 and G2251 present in the peptidyl center of the 23S rRNA [270; see also references 50, 121, and 282], but this had only very minor potential to detract from the importance of finding this CC·rRNA pairing.) Interestingly, a C74U alteration, which still should be able to wobble toward G2252, did not induce suppression. Base alterations of G2252 and G2253 in 23S rRNA, with which the tRNA CC pairs, or deletion of C1400 of 16S rRNA also influences reading frame maintenance (114, 238). In addition to the C74 mutant tRNAs suppressing a −1 frameshift mutant, they also more efficiently suppress a nonsense codon (especially UAG and UGA) if a valine codon is 5′ adjacent to the stop codon. Amino acid sequencing in these cases showed that glutamine is specified by UAG and UAA and tryptophan by UGA, so the mutant valine tRNAs reduce the ribosome's ability to discriminate against tRNAs with a first-position mismatch at the stop codon occupied A site. Thus, not only does alteration of tRNA sequence alter anticodon interaction at the opposite end of the tRNA tertiary structure, but it also affects decoding in the A site. However, these tRNA mutants can affect A-site decoding, not only from the adjacent P site but also, in certain mRNA contexts, from the E site (239), which provides a link to the next topic (see also reference 358a).
Following peptidyl transfer, the 3′ end of the deacylated tRNA moves to an intermediate state in which its amino acceptor end is in the E site and the anticodon still is in the P site. The deacylated tRNA can be in this hybrid P/E stage when the newly formed peptidyl-tRNA is still in the A/A site prior to its moving to the counterpart hybrid A/P state (214). No evidence relevant to codon-anticodon realignment has emerged from studies with frameshift mutant suppressors, but it has been considered in relation to programmed frameshifting. Even when deacylated tRNA reaches the E site, its anticodon has been proposed to be still involved in codon pairing (41). While recent structural studies are compatible with this proposal (153), they have not so far provided direct evidence for it. However, studies of the programmed autoregulatory frameshifting in decoding RF2 mRNA support the proposal in this case (22, 197, 273, 274). Alterations of 23S rRNA in the 50S part of the E site destabilizes the binding of deacylated tRNA, and such mutants also induce frameshifting (283). More pertinently for this review, mutants of the tRNA which decodes the codon present at the E site at the time of the frameshift have been tested for their effect on frameshifting. The shift site CUU UGA is highly conserved (22), and the frameshift involves peptidyl-tRNAGUGLeu (anticodon GAG) detaching from CUU and re-pairing to mRNA at UUU (underlined). In a study by Curran and colleagues (274), the tRNA mutations were in the anticodon loop, outside of the anticodon, in a derivative of tRNATrp with the anticodon 5′CUA3′, which decodes UAG. The shift cassette was a derivative of that from the RF2-coding sequence, where the codon 5′ of the CUU shift site is UAG (and the 3′ adjacent UGA stop codon is replaced by another slow-to-decode codon, CGG), i.e., UAG CUU CGG. Almost all the anticodon loop mutants increased frameshifting and did so over a fivefold range. The authors interpreted this increase as being due to destabilization of E-site codon-anticodon pairing increasing the propensity of mRNA slippage on dissociation of peptidyl-tRNA anticodon-codon pairing, supporting the relevance of the E site for framing (274). Independent data from mutant tRNA for E-site codon-anticodon pairing and its relevance to framing follow.
Ribosomal protein L1 influences strongly the ejection of deacylated tRNA from the E site (62a, 169). This protein is in close proximity to ribosomal protein L9 (Fig. 3), the presence of which restrains forward mRNA slippage (130, 131), perhaps by influencing L1-mediated efflux of the E-site tRNA. One set of experiments have been interpreted to imply that L9 senses RNA structural occupancy (normally tRNA) of the ribosomal A site before coordinating release of E-site tRNA (in accordance with allosteric occupancy of the E and A sites [41]). Though this experiment did not use frameshift mutant suppressors, another set of experiments did. The latter used a tRNAmnm5UCCGly (tRNA2Gly) C40G mutant isolated previously (129). The disruption of anticodon stem pairing permitted only a low level of −1 frameshifting at the G GGA sequence present in the S. enterica trpE91 frameshift window compared to the mnm5U34C mutant, but it was remarkably efficient in causing stop hopping at the sequence U GGA UAA GGA (131). (Consistent with other evidence that it is the mutant tRNA itself which is in the P-site tRNA mediating the detachment and re-pairing to mRNA four to six nucleotides 3′, replacing the second GGA with GGG resulted in lower activity [131].) Since the C40G mutant is especially prone to dissociate from its P-site codon, it may do the same in the E site if codon-anticodon pairing normally occurs there. On the basis that such pairing might be important for framing (273, 345), in the presence of the C40G mutant, efficient suppression of the −1 frameshift mutant trpE91 was selected on minimal medium without tryptophan in the presence of a deletion of the gene encoding ribosomal protein L9. As lack of the WT L9 constrains forward mRNA slippage, its absence permits increased WT tRNAcmo5UACVal slipping +2 from GUG to GUG in decoding the sequence G GGA GUG UGA in the short relevant “window” in the trpE91 sequence (see above and below). One of the three resulting suppressors characterized had an altered RF2; its Phe-207 was changed to Leu. (Prior genetic studies identified the peptide sequence SPF207 as functionally critical for stop codon recognition [149], and modeling based on the atomic-level structure of RF2 docking into the A site provides detailed insight into the role of Phe-207 in the process [338]). Interestingly, reintroduction of a WT L9 gene was deleterious, actually sufficiently so that a revertant was isolated and found to have its causative secondary mutation affecting the carboxy-terminal domain of L9 (17). Isolation of RF2 mutants shows that the UGA stop codon was in the A site. Thus, with the mRNA sequence G GGA GUG UGA, the location of C40G tRNAmnm5UCCGly (tRNA2Gly) was in the E site. The much stronger suppression observed when tRNAmnm5UCCGly has the C40G change than when it is WT is suggestive of the involvement of E-site codon-anticodon pairing, though follow-up work is needed.
If the interpretation of genetic data in this section pointing to E-site codon-anticodon pairing and its relevance to framing is correct, then how long will it be before structural backup is forthcoming?
CODE ORIGIN: THEORIES OF ALTERED ANTICODON LOOP STACKING AND tRNA-tRNA INTERACTION
Trinucleotide pairing, even when one of trinucleotides is in the loop of a stem-loop structure, is inherently unstable in the absence of a sophisticated ribosome environment. This has prompted thoughts about the evolutionary origins of decoding involving either more than three bases being the effective anticodon or alternative stacking that stabilizes core trinucleotide pairing. We will consider the latter first.
The involvement of most anticodon loop bases in a 3′ stack (101) is one of the hallmarks of decoding (with two bases on the 5′ side and five bases stacked on the 3′ side of the anticodon loop). This section will focus primarily on alternative forms of the 3′ stack for which, especially with certain mutant tRNAs, we believe there is strong suggestive evidence. First, however, we start with suggestions for possible switching to an alternative 5′ stack, for which there is no evidence and which is most unlikely to occur in extant ribosomes (296). A clever proposal for its possible involvement in early protein synthesis merits its inclusion, partly as a backdrop for brief consideration of whether alternative 3′ stacks are likely to have had any evolutionary significance.
The possibility of alternative 5′ stacks has been considered (286, 348), in particular for possible primordial synthesis (65). Woese (348) and Crick and colleagues (65) proposed that at the start of the ribosome cycle, the anticodon bases stacked with those further 3′ in the loop. However, they further proposed that the anticodon subsequently switched to stacking with 5′ bases. In the proposal by Crick et al. (65), two of the nonanticodon bases in both stacks also sequentially paired to mRNA and stabilized weak triplet anticodon-codon pairing in the absence of sophisticated ribosome stabilization. The elegance of the scheme was that the core codon, which was continuously involved in pairing, was three nucleotides. With the essence being a triplet code, no highly disruptive change would be involved in changing from an initial quadruplet or quintuplet code to a triplet code. Of course, requiring anticodon loop pairing of two extra mRNA bases initially on the 3′ side of the core codon and subsequently on its 5′ side would seriously restrict the number of available codons, but the initial codon set was anyway doubtless restricted. As the ribosome evolved, selection pressure could also have changed to be for stabilization of just one stack conformation, the one used now.
Before considering alternative 3′ stacking with mutant tRNAs, its possible occurrence with certain WT tRNAs with seven anticodon loop nucleotides will be considered. Even with an unperturbed balance of tRNAs, WT tRNAGGUThr (anticodon GGU) decodes CCG and CCA proline codons to cause −1 frameshifting to yield autoradiographically detectable products from E. coli extracts (counterpart reading of a GCA alanine codons by WT E. coli tRNAGCUSer requires elevated levels of the latter) (12). This involves anticodon base 35 pairing with the first codon base (and anticodon base 34 pairing with the second codon base). If the WT tRNAGGUThr (and tRNAGCUSer) in the A site are in a standard 2:5 stack, then the mRNA position must be offset (the location of the minor groove of the pairing of the first two codon bases is crucial for its normal monitoring by flipped-out 16S rRNA bases 1492 and 1493 [241]). On this standard 2:5 stack model, either two or three mRNA bases are translocated; in the latter case, the mRNA must then slip back by one base. However, the alternative possibility of a 1:6 A-site stack (i.e., six bases stacked on the 3′ side) with subsequent flipping to a 2:5 stack in the P site has been raised (13). A 1:6 stack may permit involvement of U33 of these tRNAs pairing with the third A-site codon base (termed U-33 grapple by Weiss [332]). The unstacked anticodon base, base 32, in the hypothetical 1:6 conformation spans the minor grove, and modeling has been performed (13). In contrast to this case, deductions about unusual stacking of a subset of mutant tRNAs with enlarged anticodon loops, based on their frameshift mutant suppression properties, have involved only the P site.
The anticodon of GUG-decoding tRNAcmo5UACVal is cmo5UAC. A mutant with A inserted between anticodon positions 34 and 35 (yielding the sequence AAC) competes with WT tRNA to hop from GUG to CUN in the sequence containing the Val-stop-Leu codons in the sequence GUG UAA CUN. Analysis of several combinations of takeoff and landing sites revealed that a different but overlapping anticodon is used for re-pairing to mRNA, i.e., an “anticodon shuttle” in the ribosomal P site (235). Moreover, Moore et al. (211) provided different data and reached a similar conclusion.
While the hypotheses of Woese (348) and of Crick et al. (65) do not involve a shuttle of the triplet anticodon, the studies with the eight-membered loops highlight the potential for different anticodon loop stacking conformations even with modern ribosomes. The frameshift mutant suppressors have revealed the potential for 10 extra bases (316) or even a purine at position 32 of the preceding tRNA to influence framing (211). If primordial tRNAs had eight- or nine-membered anticodon loops, it is not difficult to envisage that selective pressure for increased framing stringency generally resulted in modern tRNAs generally having seven-membered anticodon loops. The variety of products detected in E. coli with even one tRNA with an eight-membered anticodon loop (210, 211) provides an indication of the extent of the resulting multiple products. In a more primitive system, such “decoding latitude” could have facilitated escape from “barriers” to continued decoding and may occasionally have been advantageous. If proto-tRNAs with enlarged anticodon loops did function on proto-ribosomes, would unstacked 3′ anticodon loop nucleotides have sometimes spanned the major grove in a hypothetical 5′ stack?
Pondering a function for a proto-tRNA before the origin of protein synthesis led to the suggestion that proto-peptidyl-tRNA-like molecules performed some of the functions later performed by proteins (351) and that this was subsequently facilitated by direct interaction between different peptide-tRNAs (247). Coordination of the order of peptide tRNAs by a proto-mRNA could have followed and may have initially involved more than three bases of its bases pairing (83; P. V. Baranov, personal communication, 2007). Many have dismissed out of hand thoughts that primitive coding could have involved more than three bases (except possibly six) being involved in pairing because of the difficulty of retaining knowledge of what had previously been selected as the number of bases in a codon changed to three, However, some are focusing on the possibility of a nonrigid and complex variety of codon sizes prior to triplet decoding. (P. V. Baranov, personal communication) (an analogy in the Ogham alphabet used 1,500 years ago would be a mixture of the symbols for q and c and for s and n becoming like those for t and f, respectively).
Irrespective of whether proto-tRNAs had enlarged “anticodons,” it is interesting that certain WT tRNAs, e.g., the S. cerevisiae mitochondrial tRNAUAGThr gene, can also be drawn with eight bases in the anticodon loop (unless the anticodon stem has six instead of the canonical five base pairs, with the “pair” next to the loop being U·U and the loop being an unprecedented six nucleotides) (186). The same arguments suggest that the counterpart tRNA from Candida glabrata likely has an anticodon of nine nucleotides, though Santos and colleagues (208) do not favor this possibility. Current work by one of us is exploring the evolution of a natural tRNA with an eight-membered anticodon loop.
PERTURBATIONS, IMBALANCES, AND FRAMESHIFTING
Synthetic Perturbations
Primary sequence alterations and modification deficiency of tRNA causing a frameshift. (i) Alterations in the primary sequence of the amino acid acceptor stem.
Another aspect of some of the numerous interactions of rRNA and ribosomal proteins, with peptidyl-tRNA forming a grip on the latter, is that changes in the acceptor end of the peptidyl-tRNA might interfere with these interactions and thereby induce frameshifts. A G1A (see Fig. 4 for the tRNA number system used) alteration of tRNAmnm5UCCGly (sufS627 mutation) abolishes the G1-C72 base pairing in the acceptor end, and such an alteration induces −1 frameshifting (246). Also, a C70U base substitution in tRNAmnm5s2UUULys, which changes a Watson-Crick base pair to a G-U wobble pair, results in a reading frame error (315). Although the frameshifting mediated by these tRNAs is not known to occur in the P site, these changes in the acceptor end of the tRNA might change its interaction with a P-site ribosomal component (Fig. 2) and thus weaken the ribosomal grip of the peptidyl-tRNA, resulting in a shift of frame.
(ii) Alterations in the core region: TΨC and D loops.
Ribosomal protein L5 is located close to the elbow of the peptidyl-tRNA, and helix H69 is close to the hinge region in the upper part of the anticodon stem and may therefore be part of a possible ribosomal grip of the peptidyl-tRNA (Fig. 2). Alterations of tRNAmnm5UCCGly in the TΨC loop (with insertion of U expanding the loop) and base substitutions (C61U or C62A [see Fig. 4 for numbering of tRNA]) in the TΨC stem suppress the trpE91 mutation (246), and such changes in the tRNA would alter not only the structure of the TΨC loop but also its interaction with the D loop. Indeed, the C61U base substitution also induces the formation of Ψ in position 13 of the D stem, suggesting that the structure around this position is altered so as to make it a substrate for the tRNA (Ψ13) synthetase. The authors alluded to the possibility that the changes in this region of the tRNA influence its interaction with rRNA (246). Such changes in the structure may weaken the ribosomal grip of the peptidyl-tRNA and perhaps its interaction with protein L5 and/or helix H69, which are close to this region of the tRNA (170). Indeed changing the interaction between positions 18 and 55 reduces the rate of translocation of the peptidyl-tRNA, proving the importance of an intact structure of this part of the tRNA in the translocation process (250).
One class of frameshift-causing tRNAmnm5UCCGly mutants (encoded by the glyT gene) have alterations in positions G18 (to A, C, or U), G19 (to U), and C56 (to A) (131) (see Fig. 4 for numbering of tRNA). The three-dimensional structure of the tRNA is stabilized by interactions involving these bases in the D and T loops. Alterations of these interactions likely destabilize the tRNA. Indeed, C56, which is altered to A in one of the mutants, in the TΨC loop of peptidyl-tRNA interacts with ribosomal protein L5 (282).
Over 100 independent mutants of the S. enterica proM tRNAcmo5UGGPro were isolated as +1 frameshift suppressors (221). Two different parts of the tRNA were affected: the anticodon stem (positions 31, 32, 38, 40, and 43) and part of the “elbow region” composed of the D arm and its interaction with the variable arm (see Fig. 4 for explanations of positions in tRNA). The frameshift event occurs in the P site, and interestingly, the majority of these alterations in proM tRNAcmo5UGGPro are in close proximity to ribosomal components in the P site.
(iii) Alterations in the anticodon stem.
Analysis of 34 base substitutions in the anticodon stem of the E. coli amber suppressor Su7 showed that although they grossly affected the efficiency of reading the amber codon, they did not impose any frameshifts at the amber codon in the test sequence GGU UAG CGU CA (+1) or GGU UAG UCA (−1) (the underlined AGC and GUC are in the correct reading frame, and the UAG codon is in the zero frame) (79). The sequence allows a frameshift at GGU to GUU by the WT peptidyl-tRNACCGGly, assuming that UAG is in the A site. Indeed, an excellent correlation was observed between decreasing frameshifting and increasing efficiency of the amber suppressor, consistent with a frameshifting event in the P site by tRNACCGGly. However, it has not been established that frameshifting occurs at the GGU (Gly) codons, and it is unclear if these results support a defective ribosomal grip of the peptidyl-tRNA (their effects when in the E site are described above) or rather that slow entry by the mutant tRNA to the A site stimulates frameshifting in the P site even by WT tRNACCGGly.
Several S. cerevisiae SUF8 alleles with various base substitutions at the last base pair in the anticodon stem (31-39, i.e., the base pair next to the anticodon loop [Fig. 4]) of tRNAncm5UGGPro induce frameshifts of the his4-713 allele, which contains a CCAA sequence within its frameshift window (200). Suppression was observed only when base pairing between positions 31 and 39 was lost; this creates an extended anticodon loop (i.e., base substitutions that do not disrupt base pairing did not induce frameshifting). The location of SUF8-encoded tRNA in the ribosome when it induces the frameshift event was not tested, but for another yeast proline tRNA with an expanded anticodon loop (SUF10) a P-site location was established (259). Since these base substitutions clearly change the conformation of the tRNA and since they also influence the processing of the tRNA, they may also affect potential interactions between the peptidyl-tRNA and the P-site ribosomal components.
A yeast mitochondrial tRNAUGASer altered at position 42 (second base pair in the anticodon stem) from C to U changes a Watson-Crick base pair to a wobble G-U base pair. In addition it also abolishes the formation of Ψ at position 27. Such changes confer the ability to suppress a +1 frameshift mutation in the oxy1 gene of yeast mitochondria (143). The WT form of this tRNA was suggested to read all four serine UCN codons and the mutated tRNAUGASer to cause frameshifting by decoding four bases. Later it was suggested that the defective tRNAUGASer induces frameshifting either by decoding the UCC codon in the A site, poorly allowing the peptidyl-tRNA to shift frame, or by allowing a competing proline tRNA to misread the UCC codon and in turn inducing a shift in frame by the peptidyl-tRNA (269). Interestingly, an altered tRNAGGGPro with a base substitution at the first base pair in the anticodon stem (i.e., next to the one altered in yeast mitochondrial tRNAUGASer) also induces frameshifting by allowing the third-position-mismatched tRNAcmo5UGGPro to read the CCC codon, and following a normal translocation, this WT third-position-mismatched tRNA shifts frame in the P site (258, 259, 295). In fact, several other alterations in tRNAGGGPro, including the classical Salmonella frameshift suppressor sufB, induce +1 frameshifting by such a mechanism. Thus, some anticodon stem alteration may induce frameshifting by causing a slow decoding of the A-site codon, allowing the WT peptidyl-tRNA to shift frame.
A section above, which dealt with the E site, includes usage of a mutant GGA-decoding tRNAmnm5UCCGly with the anticodon stem substitution C40G. The change results in a G-G purine clash in the anticodon stem and lack of the modification (mnm5U34) normally present in the tRNAmnm5UCCGly (129). A1339 of 16S rRNA reaches the G30-C40 base pair in the minor groove of the peptidyl-tRNA (282). A change from a GC base pair to a G-G mismatch most likely changes the interaction between the tRNA and the rRNA. In addition, lack of the mnm5 or the mcm5 group of the wobble nucleoside reduces the cognate decoding of GGA (38, 320). While the relationship of the inferred reduced ribosomal grip of the mutant tRNAmnm5UCCGly to its dramatic increase in stop-hopping ability remains to be clarified, it is easier to explain its greatly reduced ability to act as peptidyl-tRNA for programmed 50-nucleotide translational bypassing (unpublished results). (Although the substitution destabilizes the tRNA, resulting in a smaller amount of it compared to WT tRNAmnm5UCCGly, the programmed bypassing monitors ribosomes that have already translated codon 46 [GGA], and therefore its reduced level should not interfere with bypassing.)
(iv) Some missense and nonsense suppressor tRNAs also suppress frameshift mutations.
Isolated missense and nonsense suppressors, which have an alteration in the anticodon region, can also suppress frameshift mutations (315). One of the missense suppressors was the first identified case where a triplet anticodon was offset, i.e., not in the standard position (217). Among 22 different missense and nonsense suppressors tested, some have an eight-membered anticodon loop and some have changes in the anticodon preserving the normal seven-membered anticodon loop. Several mediate suppression of +1 frameshift mutations, demonstrating no simple correlation between frameshifting and seven- or eight-membered anticodon loops (315). However, the sequence of the frameshift product was not established.
It should also be remembered that several mutants isolated as frameshift suppressors are either known or likely to suppress nonsense or missense mutations; e.g., the hopR tRNAcmo5UACVal mutant inserts one amino acid (Val) at the sequence GUG-UAA-GUU (235) and in doing so circumvents the stop codon.
(v) Frameshifting induced by modification deficiency.
The importance of tRNA modification for reading frame maintenance was first revealed when it was discovered that lack of m1G37 induces frameshifting at CCCN codons (40). The involvement of m1G37 in reading frame maintenance has been discussed above, but a shift by the m1G37-deficient peptidyl-tRNAPro is induced by its altered interaction with the ribosomal components in the P site.
There are two principally different ways that tRNA modification deficiency, as well as alterations in the primary sequence of the tRNA, can influence reading frame maintenance (320). Lack of a modified nucleoside in tRNA may affect reading frame maintenance either by altering the rate at which the ternary complex binds to the ribosomal A site or by altering the interaction between the ribosomal P site and peptidyl-tRNA. Figure 6 shows a model predicting how a hypomodified tRNA or an otherwise defective tRNA induces frameshifting. In the first case (Fig. 6A), a hypomodified cognate tRNA enters the ribosomal A site sufficiently slowly that a WT third-position-mismatched tRNA can out-compete it and be accepted to the A site (A-site effect by hypomodified tRNA). Following a normal three-nucleotide translocation, this third-position-mismatched peptidyl-tRNA is nonoptimal in the P site, which results in a +1 frameshift (28, 122, 259, 304, 320). In the second case (Fig. 6B), the hypomodification causes a slow entry of the hypomodified cognate tRNA to the A site and thereby induces a pause that allows the WT cognate peptidyl-tRNA to slip to the +1 frame (A-site effect by hypomodifed tRNA). Alternatively (Fig. 6C), the defective or the hypomodified cognate tRNA does not affect the A-site selection step, but rather the undermodification disrupts the interaction between the ribosomal P site and the peptidyl-tRNA after the translocation step. Modification deficiency changes the structure of the tRNA and thereby weakens its interactions with ribosomal components in the P site, similar to the weakened interaction by a third-position-mismatched tRNA. This results in an increased frequency of frameshifting (P-site effect by undermodified tRNA). Of course, undermodfied tRNAs may cause frameshifting by mediating both A- and P-site effects.
FIG. 6.
The dual-error model for frameshifting. Defects in tRNA may induce frameshifting in three different ways. (A) The defective tRNA is slow in entering the A site, allowing a third-position-mismatched tRNA to decode the A-site codon (first error). After a normal three-nucleotide translocation to the P site, the third-position-mismatched tRNA is prone to slip into an overlapping reading frame (second error). (B) The defective tRNA is slow in entering the A site (first error), providing a pause when the P-site tRNA may slip (second error). (C) The defective tRNA is not excluded by the ribosomal A site and decodes the codon in the A site (first error), but once it has been translocated into the P site, it may slip on the mRNA (second error). “Defective” can indicate either alterations in the primary sequence or hypo- or hypermodification of the tRNA. To make the figure easier to read, no tRNA has been depicted in the E site, although in all these cases when a frameshift occurs in the P site (lower part of the figure), it is likely that the E site is occupied (see text). (Adapted from reference 37.)
Increased +1 frameshifting was observed for several tRNA modification-deficient E. coli or S. enterica mutants (mnmA, mnmE, tgt, truA [hisT], trmD, miaA, miaB, and miaC mutants) (320). Generally, if an A-site effect is observed, a P-site effect is also observed by the same tRNA (e.g., mnm5s2U34 in tRNALys, ms2io6A37 in tRNATyr, m1G37 in tRNAPro, and m1G37 in tRNAArg). In two cases (tRNAPhe deficient in ms2io6A37 and tRNAGln deficient in mnm5s2U34), no A-site effect occurs although there is a strong P-site effect. Lack of the mnm5 modification of mnm5s2U34 of tRNAmnm5s2UUCGlu induces a +2 (or −1) frameshift by mechanisms essentially as described in Fig. 6, but the slippage in the P site is either two nucleotides forward or one nucleotide backward (47). Also, lack of ms2i6A37 of tRNAGAAPhe induces frameshifting at the sequence UUU-YNN, which is rare in E. coli genes, and this frameshifting increases in the stationary phase of growth (280). Thus, although these modified nucleosides vary greatly in their chemical structures, their presence in different tRNAs, and their locations within tRNAs, they all improve reading frame maintenance by promoting efficient A-site selection, preventing peptidyl-tRNA slippage, or both.
According to the model (Fig. 6), the peptidyl-tRNA should also be able to slip in the −1 direction. However, the above-mentioned bacterial mutants defective in tRNA modification that induce +1 frameshifting rarely induce −1 frameshifting at several heptameric sequences (U-UUU-UUA, G-UUU-UAU, A-AAA-AAC, and G-GGU-UUA [321]). In one case, the presence of the mnm5 or the s2 modification of the mnm5s2U34 in tRNALys increases the frequency of −1 frameshifting at the sequence A-AAA-AAC. Similarly, the presence of either the mnm5 or the s2 modification in tRNAmnm5s2UUULys increases −1 frameshifting at the sequence NNA-AAG, when NNA was GCA, GUA, or CCA (188), whereas the mnm5 modification but not the s2 modification of mnm5s2U34 in tRNAmnm5s2UUULys decreases frameshifting at the slippery site U-UUA-AAA (48). Q34 in tRNAQUUAsn has only a slight influence on −1 frameshifting as monitored in E. coli mutants defective in the synthesis of Q (48). Thus, in contrast to the pivotal role of many modified nucleosides in preventing +1 frameshifting, the presence of modified nucleosides in bacteria has only a marginal or no influence on −1 frameshifting.
As mentioned above, lack of the s2 or the mnm5 (mcm5 in eukaryotes) group influences reading frame maintenance by either inducing +1 or −1 frameshifts or decreasing −1 frameshifts. The s2 group is present in tRNAs from all organisms reading codons of the general type NAR, i.e., tRNAs specific for Gln, Lys, and Glu. These tRNAs have the anticodon sequence 5′-U33-U34-U35-N36-Pu37-3′. As the stacking potential of U is poor (319), the unmodified anticodon loops of these tRNAs are inherently unstable and therefore unstructured. The Lys-tRNA is unique in this sense, since its anticodon is composed of a stretch of three Us. To structure the anticodons of these tRNAs, and especially the Lys-tRNA, the modifications, such as s2 (in all organisms [except Mycoplasma] and organelles), mnm5 (in Bacteria, Archaea, and mitochondria), mcm5 (in Eukarya) and t6A37 (only in Lys-tRNA but present in all organisms), are pivotal (141, 204, 287, 303).The ubiquitously present s2 group improves the stacking ability of U (141, 204, 287, 303), and its presence in this set of tRNAs in various organisms is caused by convergent evolution, since two unrelated proteins are responsible for its synthesis in the three domains Eukarya, Archaea, and Bacteria (38). Thus, the evolutionary driving force for the presence of this type of modification (xm5s2U) has provided a very strong explanation of its ubiquitous presence in this set of tRNAs from all organisms, except Mycoplasma, and in organelles. Indeed, viability of yeast is dependent on the presence of mcm5s2U34 in the wobble position of the Gln-tRNA and especially the Lys-tRNA (38).
The YbbB protein has been suggested to catalyze exchange of the sulfur of mnm5s2U34 in tRNA with selenium, thereby forming mnm5se2U34 (350). A dominant (“gain-of-function”) mutation (sufY204), resulting in an amino acid substitution in the protein YbbB, suppresses several +1 frameshift mutations (60). The altered YbbB possesses a novel catalytic activity resulting in the addition of a geranyl group (or an isomer of it, C10H17, with a molecular weight of 137) to the wobble nucleoside cmnm5s2U34 of tRNAcmnm5s2UUGGln and most likely also to the other mnm5s2U34-containing tRNAs specific for Lys and Glu, i.e., the same modified nucleoside which can be changed to the selenium derivative mnm5se2U34 in the same tRNAs. However, this novel activity mediated by amino acid substitution of YbbB is not dependent on selenation, since it still occurs in a selD mutant which is blocked in the synthesis of the donor of selenium (selenophosphate) required for the selenation reaction. Such altered tRNAcmnm5s2UUGGln is inefficiently aminoacylated, and it is questionable whether this altered tRNA with such a large extra modification in its anticodon is able to enter the A-site CAA codon in the sequence -CCC-CAA-UAG (the codons are in the zero frame; the +1 frameshift insertion is underlined). Therefore, the modification causes a low concentration of charged tRNAcmnm5s2UUGGln and so induces an extended pause that allows third-position-mismatched peptidyl-tRNAcmo5UGGPro (not the cognate tRNAGGGPro, since lack of cmo5U34 reduces the frequency of frameshifting [Fig. 6A]) to shift to the +1 frame according to the model shown in Fig. 6A. These results demonstrate that hypermodification of a tRNA can also induce frameshifting and reinforce the viewpoint that modification of tRNAs generally optimizes tRNA for standard translation.
The absence of Ψ in the anticodon stem of bacterial Leu-tRNAs, as in the truA (hisT) mutant, increases +1 frameshifting by an A-site effect at three of the four leucine codons tested and by a P-site effect on one leucine codon (CUA). However, such deficiency did not influence frameshifting at CCC (Pro) or CAU (His) codons (320). In yeast, Ψ39 deficiency in tRNAUAGLeu decreases +1 frameshifting at the slippery Ty1 CUU AGG sequence (179). Thus, Ψ deficiency in the anticodon stem in some bacterial tRNAs increases +1 frameshifting, whereas in other tRNAs it does not influence reading frame maintenance. Thus, the influence on reading frame maintenance by Ψ in the anticodon stem is dependent on the tRNA, frameshifting site, and organism.
One of the most complex modified nucleosides present in tRNA is wyebutosine (yW37), which is present in yeast tRNAGmAAPhe. The synthesis of yW37 requires several biosynthetic steps to convert the encoded G37 in the tRNA. The first step is the conversion of G37 to m1G37 catalyzed by Trm5p (39) followed by the action of four additional enzymes (TYW1p to TYW4p) (324). Thus, the biosynthetic pathway to modified G37 of tRNAGmAAPhe to yW37 is as follows: G37 (Trm5) m1G (TYW1p) imG-14 (TYW2p) yW-86 (TYW3p-TYW4p) yW37 (identified intermediates are indicated, and the enzymes involved in the conversion are indicated within parentheses between the substrate and the product). Mutations in the gene TRM5, which results in the presence of unmodified G37 in yeast tRNAGmAAPhe, does not influence −1 frameshifting at the human immunodeficiency virus sequence U-UUU-UUA (321). However, at the S. cerevisiae virus La sequence G-GGU-UUU, the presence at the A site of hypomodified tRNAGmAAPhe containing m1G37 instead of yW, as in a tyw1 mutant, increases −1 frameshifting. The presence of imG-14, as in a tyw2 mutant, also increases frameshifting but to a lesser extent than the presence of m1G37 (325). Thus, in yeast, hypomodification of tRNAGmAAPhe in the A site at the S. cerevisiae virus La heptameric sequence results in increased −1 frameshifting.
The heptameric sequences in many retroviral genomes at which programmed −1 frameshifting generates its GagPol, or GagProPol, product have the general sequence X XXY YYZ. The initial model for the realignment to the −1 frame (150) was followed by several others. The A-site codon (YYZ) is often AAC, which is decoded by the Q34-containing tRNAAsn, or UUU, which is decoded by the yW-containing tRNAGAAPhe (125). In several cases, both A- and P-site codons are read by tRNAs having such bulky modifications at either position 34 or 37. Upon virus infection, tRNAGAAPhe and tRNAAsn become yW and Q deficient, respectively (124). It was therefore suggested that the “shifty” tRNAs involved in the simultaneous −1 frameshifting are hypomodified isoacceptors and that such hypomodification is a prerequisite for an efficient −1 frameshifting. Indeed, the results mentioned above demonstrate in vivo in yeast that yW37 deficiency of tRNAGmAAPhe induces frameshifting, thereby supporting such a suggestion. Also in oocytes, deficiency of yW37 induces −1 frameshifting at the sequence A AAA AAU (55). Moreover, in vitro, tRNAGAAPhe containing m1G37 instead of yW37 slightly stimulates −1 frameshifting at the human immunodeficiency virus sequence U-UUU-UUU (56), and Q deficiency of tRNAAsn does so at the A AAA AAC sequence (54). However, at the latter sequence in oocytes and at the coronavirus infectious bronchitis virus signal U UUA AAC, Q34 deficiency in tRNAAsn does not influence −1 frameshifting either in vivo or in vitro (55, 196). Apparently, −1 frameshifting at heptameric sequences is dependent on tRNA modification, although it is dependent on which protein synthesizing system is used to monitor it as well as on the heptameric sequence. Still, an adequate modification(s) of the tRNA influences −1 frameshifting and thereby may play a key role in setting the level of frameshifting important for optimal gene expression.
These results demonstrate that hypo- or hypermodification induces framing errors and reinforce the viewpoint that modification optimizes tRNA to decode mRNA efficiently and accurately. Frameshifting can be induced by modification deficiency at two important steps in translation: at the A-site selection step and while the tRNA is residing in the P site as peptidyl-tRNA (Fig. 6). The former effect is consistent with the suggestion that the importance of tRNA modification is partly in equalizing the affinities of the various ternary complexes for binding to the codon in the A site (91). Although deficiency of some modified nucleosides (e.g., k2C34, I34, mcm5s2U34, or m1G37) exerts a strong effect on viability, some of them induce no or only a mild, phenotypic growth defect besides inducing frameshift errors. This is not intended to imply that selection does not in parallel operate to promote a tRNA set optimal for specific frameshifting that is utilized for gene expression. An example may be having only one lysine isoacceptor in E. coli but two in certain other bacteria. Poor pairing of E. coli tRNAmnm5s2UUULys with AAG in the sequence A AAA AAG, which is present in the dnaX gene, makes it highly shift prone (314, 335). This shiftiness is dependent on the presence of mnm5s2U34 in tRNAmnm5s2UUULys when it decodes either the P-site codon or the A-site codon (188, 321). Arguably, one lysine tRNA and its modification have coevolved to adjust a proper frequency of frameshifting at A AAG. Though the frameshift event in decoding E. coli dnaX is not essential (42), the utilization of a different mechanism to generate two counterpart polymerase products from the homologue in a thermophile (175) suggests that it is selectively advantageous.
Ribosomal mutants.
The section dealing with ribosomal sites above includes information about some specific effects of certain rRNA mutants. rRNA or rRNA modification mutants with decreased fidelity, including those with restricted frameshifting (342) and with elevated frameshifting, will not be discussed here, since this topic will be covered in an upcoming review by J. D. Dinman and M. O'Connor.
The bacterial ribosomal protein L9 is located close to the E site and has been visualized in two different conformations. In one the N-terminal globular domain is tethered to the L1 domain of the ribosome, and the rest of L9 projects outward from the ribosome (Fig. 3). In the other, both the C- and N-terminal RNA-binding ends are tethered to the rest of the ribosome, and the central alpha-helical region is bulged out. Among the selections which yielded mutants of L9 were isolations of frameshift suppressors (128, 180; C. Johnston, cited in reference 17). As deduced from the phenotype of L9 mutants, its function is to prevent forward mRNA slippage.
The C-terminal end of ribosomal protein S9 penetrates the ribosome like a tentacle, and the two last amino acids make contact with the 5′-phosphate of nucleotide 32 (R130) and the 5′-phosphates of positions 33 and 34 (K129) of peptidyl-tRNA (Fig. 2) (282). Accordingly, ribosomal protein S9 may be a functional part of the ribosomal P site to maintain the reading frame by securing a grip of the peptidyl-tRNA. A selection for extragenic suppressors of a +1 frameshift mutation in the S. enterica his operon resulted in the isolation of mutants with a truncation of the C-terminal end of S9 (221). In addition, the two last amino acids (K129 and R130) were individually replaced with alanine and also found to suppress the +1 frameshift mutation. These results are consistent with ribosomal protein S9 being part of the ribosomal grip of the peptidyl-tRNA and pivotal for reading frame maintenance. Combinations of alterations in ribosomal protein S9 with specific alterations in the proM tRNAcmo5UGGPro (see above) suggest that an interaction occurs between the C-terminal end of ribosomal protein S9 and position 32 of the peptidyl-tRNA. Combined, these results suggest that the “ribosomal grip” of the peptidyl-tRNA is pivotal for reading frame maintenance (221).
A separate class of frameshift mutant suppressors have been identified in yeast and shown to be mutants of ribosomal protein S3 (127).
Certain restrictive alleles of ribosomal protein S12 reduced −1 frameshifting at GGA mediated by overexpressed WT tRNACCCGly (tRNA1Gly) (and abolished its suppression of trpE91, although a nonrestrictive allele enhanced it) (227). Some restrictive alleles also affect some cases of frameshift mutant leakiness (10). Potentially different effects of impaired A-site selection on different types of frameshifting may be paralleled by contrasting effects at different sites of compounds that affect the process (343).
Elongation and release factors.
Various mutants of EF-Tu/EF-1α suppress some +1 and −1 frameshift mutations (84, 95, 142, 268, 272, 318, 323). In most of these reports, the experimental work did not indicate a specific mechanism by which the altered EF-Tu/EF-1α mediates the compensatory frameshifting. An explanation for such frameshifting would be a slow decoding of the A-site codon, allowing higher probability of P-site realignment. Delayed accommodation of the ternary complex (EF-Tu*aa-tRNA*GTP) from the A/T site to the A/A site and/or initial acceptance to the A/T site may also be expected to similarly permit increased P-site realignment. Mutation of EF-Tu/EF-1α may cause such a delay and thereby induce frameshifting. However, this simple model is unlikely, since the mutated elongation factor did not appear to prolong the translational pause (95). Specific altered forms of EF-Tu/EF-1α indeed induce frameshift errors, although the mechanism behind this phenomenon is not clear.
Knowing that EF-G stimulates translocation, one would expect that alteration in this protein would induce errors in the size of the translocation step and thereby frameshifting. From this viewpoint it is surprising that so few mutant EF-Gs have been characterized (193). However, this deficiency of altered EF-G mutants inducing frameshifting is consistent with EF-G having a stimulatory rather than a qualitative function in translocation and also with the fact that it has no role in reading frame maintenance.
Stop codons are slowly decoded, and defective or reduced amounts of release factors exacerbate this effect, thereby increasing the propensity of frameshifting at the 5′ adjacent P-site codon. This is evident with altered levels of WT release factors (2, 64), with certain mutant release factors (reviewed in reference 17) or when a WT release factor undergoes evolutionary adaptation (164). One class of the yeast frameshift mutant suppressors are in a gene (346) which is now known to encode eRF3.
Imposed changes in tRNA levels.
In E. coli, the tRNA population is correlated to codon usage, and this relationship supports a maximal growth rate (29, 87). In Salmonella, Mycoplasma, and Saccharomyces, there is also a positive correlation between tRNA abundance and codon usage (144-147, 156, 284, 353). Apparently the concentration of tRNA is related to the need for obtaining smooth and efficient translation without ribosomal stalling. The elongation rate in bacteria varies with the growth rate from about 12 to 21 amino acids per second at 37°C, and the synthesis of most of the tRNAs is regulated in a growth rate-dependent manner. However, a few tRNA species do not show such regulation (87), suggesting that under some conditions there might be an imbalance between tRNA concentration and codons to be read. The amount of EF-Tu also varies positively with growth rate, as do other components of the translation apparatus, most prominently ribosomal components. Thus, the various components of the translation apparatus are coregulated to maximize growth. Any disturbance in the balance between them interferes with translation and may induce various errors.
Intuitively, one would assume that each codon should be translated at the same rate, but this seems not to be true. Early work (251, 322) found that the rate of translation is dependent on the tRNA concentration and codon choice. Later, Sörensen et al. (288) showed that a major determinant of difference in translation rate is codon usage. Two synonymous codons, GAA and GAG, which are translated by the same tRNA in E. coli, are translated with a 3.4-fold difference in rate. Since the same tRNA reads both codons, the difference in translation rate is not caused by different tRNA concentrations but is caused by the difference in reading the synonymous codons (289). As expected, GAA, which is translated faster than GAG, is also used in highly expressed mRNAs, a correlation noted earlier (288, 322), although there is evidence that this relationship may not be general (43). The use of rare codons appears to influence the rate of translation (see, e.g., references 168 and 322) and protein folding (352). Interestingly, certain combinations of single-nucleotide polymorphisms, which do not change the sequence of the encoded polypeptide, result in different activity of a eukaryotic transport protein. This phenomenon is aggravated at higher mRNA levels and thus may be dependent on relative tRNA depletion (163). Also, in bacteria silent mutations influence folding (63). Clearly, the elongation rate is sensitive to codon usage and to the concentration of various components of the translation apparatus. Disturbances in ratios between the various components and the translation apparatus may induce aberrant folding of proteins and translation errors, including those of framing.
Perturbations in the balance of certain aminoacyl-tRNAs are especially prone to lead to frameshifting. One way in which such imbalances can occur is by specific amino acid starvation and consequent ribosome stalling. Such conditions induce frameshifting (both +1 and −1) and bypassing (106, 107, 331, 336). A high level of −1 frameshifting occurs at the sequence U-UUC-AUA by limiting Ile-tRNA reading the AUA codon (21). Protein sequencing and other work revealed that the frameshift event occurs by peptidyl-tRNAPhe shifting from pairing with UUC to U-UU. Translation continued in the −1 frame with tRNAHis decoding C-AU. At the sequence UUU AUA-U, the same limitation causes a shift by peptidyl-tRNAPhe from UUU to UUA, allowing tRNATyr to read the next +1 frame U-AU codon. The frequency of bypassing (defined as an event involving re-pairing to mRNA at a nonoverlapping codon [i.e., frame independent]) is dependent on the “lightness” of the peptidyl tRNA at the “takeoff” site (105). Tests of almost all triplets as “takeoff” sites revealed that elevated bypassing is correlated with a high frequency of A·U base pairs and with codons ending with G rather than C. Arg-tRNA with inosine as wobble nucleoside and reading CGU, CGC, and CGA codons bypasses more frequently from the CGA codon, consistent with poor decoding of A by I34 (78). Since not only limitation of Ile-tRNA causes bypass, the ability to bypass upon limitation of an amino acid seems general, provided that reasonable takeoff and landing sites are available in the mRNA sequence. However, this kind of bypassing also occurs in logarithmically growing cells without imposing any aminoacyl-tRNA limitation (190). In the studies described above, the values directly measured reflect the combination of anticodon dissociation and re-pairing to mRNA, whereas in a complementary study the values reflect only re-pairing (51).
Alteration of the aminoacyl-tRNA balance can also be achieved by adding an excess of tRNA to an in vitro protein-synthesizing system. When this was performed with E. coli, only 2 out of the 33 tRNAs tested gave dramatically elevated frameshifting, and both mediated frameshifting at noncognate codons. Addition of a large excess of purified tRNAGCUSer (also called tRNA3Ser, which decodes AGU/C) resulted in −1 frameshifting at alanine codons, at least at GCA, and could be inhibited by also adding excess cognate alanine tRNA (12, 81). It was proposed that this tRNA formed Watson-Crick base pairing between the 5′ two first bases in the anticodon (C35 and G34) and the 5′ two codon bases GC. If only doublet pairing occurred, this could result in a −1 frameshift (49) and be an A-site event. In this scenario, the pairing involved is fully matched, but the anticodon is offset. However, three mRNAs bases may initially move to the P site, perhaps especially if anticodon base 33 pairs with or occludes the third codon base and a rearrangement in the P site mediates frameshifting (discussed in reference 13). (In any event, the nature of the monitoring [241] of codon-anticodon interaction may be distinctive.) Interestingly, corresponding −1 frameshifting was also observed in extracts with an unperturbed tRNA balance due to tRNAGGUThr (also called tRNA3Thr, which decodes ACU/C) decoding CCG and probably CCA proline codons (12, 81).
As introduced above, overexpression of tRNACCCGly (tRNA1Gly) in E. coli results in it now effectively competing with tRNAmnm5UCCGly (tRNA2Gly) for reading GGA, and it causes frameshifting at this codon (227). Increasing the endogenous synthesis of the cognate GGA decoding tRNA (tRNAmnm5UCCGly) abolishes frameshifting (227), as the balance is restored.
Similarly, overexpression of tRNAcmo5UGGPro induces +1 frameshifting at the third-position-mismatched CCC codon, and co-overexpression of the cognate tRNAGGGPro abolishes frameshifting. It was proposed that the third-position-mismatched tRNAcmo5UGGPro out-competes the cognate tRNA, and following a normal triplet translocation, it slipped forward one base provided that the third-position-mismatched tRNAcmo5UGGPro could efficiently base pair with the next codon (229), consistent with an observation based on depletion of the cognate tRNAGGGPro (259).
An unbalanced tRNA population may also occur if for some reason a tRNA species is not made or is rapidly degraded. If the cognate tRNAGGGPro, which reads the CCC codon, is altered or not synthesized, the third-position-mismatched tRNAcmo5UGGPro for this codon can be accepted, and when translocated to the P site, it induces a +1 frameshift in a manner similar to overexpression of the same third-position-mismatched tRNA (259). Another way to relatively deplete a tRNA species is to overproduce an mRNA which contains a rare codon, as described in the next section.
In summary, there can be several ways in which the components of the translation apparatus are not in balance, which in turn may cause nonstandard framing.
Imposed changes in mRNA levels.
When two consecutive minor E. coli codons, especially AGG-AGG or AGA-AGA, occur in highly overexpressed mRNA, a high level of frameshifting ensues (291). An increasing concentration of the cognate tRNA reduces frameshifting, suggesting that the cause of frameshifting is its limited amount (291). The explanation proposed (291) is that slow decoding of the first codon of the pair and sequestration of the limiting amount of sparse cognate tRNA leads to a stall when the second codon is in the A site. This pause increases the possibility for peptidyl-tRNA anticodon dissociation from mRNA and subsequent realignment in a new frame. A subsequent analysis showed that when the same sequences were expressed from a single copy of the gene on the chromosome, the level of frameshifting was three- to fourfold lower than when they were expressed from a multicopy plasmid. Thus, the level of frameshifting with these rare codons is dependent on the level of mRNA (117). In E. coli, AGG-AGG and AGA-AGA are confined to mRNAs that are not highly expressed, and the load of aberrant proteins is modest (117). Though unknown at present, the interesting possibility remains that the frameshifting which occurs at some such sequence pairs generates a beneficial transframe product, perhaps especially under starvation conditions. However, limitation of specific aminoacyl-tRNA not only can lead to frameshifting, but bypassing can also ensue. This was first detected with high-level heterologous expression of a mammalian gene in E. coli where there was a high demand for a tRNA isoacceptor which is normally present in small amounts (157).
Overexpression of specific mRNAs may also lead to a shortage of some specific aminoacyl-tRNA(s) relative to the amount of codons to be read. This may in turn induce mistranslation, as has been observed for incorporation of Lys instead of Arg at the Arg codons AGG and AGA (see, e.g., references 52, 171, and 281). When such a tRNA is translocated into the P site, it may induce frameshifting according to the model presented in Fig. 6C; i.e., a nonoptimal tRNA in the P site may induce P-site frameshifting. Since misreading may in some cases approach 50% (171), a substantial frameshifting may occur during such conditions.
relA, uncharged tRNA, and frameshifting.
Synthesis of ppGpp under starvation condition is dependent on the relA locus (58). Starvation of an amino acid elicits frameshifting, both −1 and +1 (107). Interestingly, the differential rate of frameshifting, which is constant in most cases, is larger in a relA mutant than in a WT (relA+) background, suggesting that some stimulatory factor for frameshifting is present or that destruction of an inhibitory factor occurs upon starvation in the relA mutant. With one of three different constructs and starvation for the amino acid required for acylation of the cognate A-site tRNA (Ile starvation), the differential rate of frameshifting increased during the course of starvation, suggesting the accumulation of a factor(s) stimulating frameshifting occurred (198). Since ppGpp deficiency elicits uncontrolled synthesis of tRNA, undermodified tRNA accumulates under such a condition (reviewed in reference 36). As such tRNA induces frameshifting (see above), it was suggested that the stimulatory factor in this case was undermodified tRNA (198). Although the molecular mechanism for the relA-mediated stimulation of frameshifting is not known either in this case or at the more general lower level, these observations may be relevant for various regulatory features depending on frameshifting (e.g., for transposition of insertion sequence [IS] elements whose transposase requires frameshifting for its synthesis).
Normally, the aminoacylated tRNA enters the A site associated with EF-Tu, but unacylated tRNA can also enter the A site, thus without being associated with EF-Tu (58). Though not established, there is evidence that when the A site is vacant for a prolonged time, for instance due to amino acid starvation, ribosomes are in a more open conformation (240). Such “hungry” A sites (using Jon Gallant's wording) may be filled by a cognate unacylated tRNA. When they are unacylated, and thus not associated with EF-Tu, the structures of the various tRNAs are more different than when the tRNAs are aminoacylated and associated with EF-Tu (174). When an unacylated tRNA is bound to the A-site codon, it will block possible +1 frameshifting by the peptidyl-tRNA. Since it is likely that the bindings of various unacylated tRNAs are very different (174), the observed different frequencies of frameshifting may be correlated with the ability of unacylated tRNA to bind to the A site. However, the only relevant data are indirect and may be explicable in other ways (11). In principle, by analogy with the effect of a stop codon stimulating −1 frameshifting (112, 334), binding of unacylated tRNA in the A site may stimulate −1 frameshifting more effectively than a vacant A site, since the time required for replacing such a tRNA with an acylated tRNA associated with EF-Tu is longer. It may not be a coincidence that all IS elements that utilize frameshifting for expression of their transposase involve −1 rather than +1 frameshifting. IS element transposition may be especially advantageous at the onset of “hard” times (160, 242). Whether binding of uncharged tRNA to the A site of viral mRNA −1 frameshift sites occurs at the late stage of infection with viruses whose decoding comes to dominate the host translation apparatus merits investigation. (Synthesis of murine leukemia virus Pol protein requires stop codon readthrough at the 3′ end of its gag gene rather than frameshifting but has features suggestive of a regulatory mechanism which causes elevated levels of readthrough at the later stages of infection [248]). Is an equivalent end result achieved by retroviruses that utilize frameshifting by exploitation of uncharged tRNA?
Natural Perturbations
Stationary-phase-induced frameshifting: a possible prelude to effects of other physiological changes relevant to frameshifting.
Perturbation of the balance of cellular components occurs as a bacterial culture enters stationary phase. Correspondingly, some frameshift mutations are suppressed more efficiently at the onset of, and during, stationary phase. The E. coli argI gene encodes ornithine transcarbamylase, and its third codon, UUU (Phe), is directly followed by a UAU (Tyr) codon (100). A surprisingly high level of +1 frameshifting (3%) occurs at the UUU codon, and this frequency increases even further, to 16%, at high cell density (at the onset of stationary phase or later). A similar level of frameshifting is also observed when the UUU (Phe) codon is followed by a CAU (His) codon but not when it is followed by a AAU or when the UUU (Phe) codon is changed to the other Phe codon, UUC. Fu and Parker suggested that the tRNAGAAPhe detaches from mRNA and re-pairs to it in the +1 frame (100). However, what causes this slip either in logarithmically growing cells or at a high cell density was not elucidated. In a separate study UUU was positioned in the RF2 programmed frameshifting site (replacing the WT CUU), and, as expected from earlier work, frameshifting was sensitive to a pause at the A site. The frequency of frameshifting increased at high cell density (280) in accordance with the first suggestion for frameshifting in the argI system. This increase in frameshifting is also exaggerated by lack of ms2i6A37 in tRNAGAAPhe. One reason for the increased frameshifting observed at high cell density may be that the ratio between acylated and unacylated tRNA changes at such a growth phase or that some tRNAs synthesized during this growth phase become undermodified. Transiently increased frameshifting at an exceptionally slippery sequence (nine Us in a row) was observed at the onset of stationary phase (339), though standard translation of mRNA with extra Us due to transcription slippage (175) doubtless accounts for a substantial portion of the product.
An exceptionally large increase (30-fold) of E. coli −1 frameshifting at a U-UUC-AAG site has been detected as cells enter the stationary phase (20). The sequence of the protein revealed that the frameshifting occurred either by the pairing of two tRNAs (Lys [AAG] and Phe [UUC]) slipping −1 or by slippage of the Phe-tRNA from UUC to U-UU followed by binding of Gln-tRNA (CAA) to the A site.
The findings just described are all from bacterial systems, and in all cases the frameshift mutation suppressed was plasmid borne. The cause of the increased +1 and −1 frameshifting is not known, though a changed ratio in key tRNAs of acylated to unacylated forms and the generally slower translation are likely relevant (20).
A contrasting result has been obtained in S. cerevisiae for the programmed frameshifting event required for synthesis of the Ty1 transposase (299). Upon entering the stationary phase, a gradual decrease of +1 frameshifting was observed, i.e., opposite to the observations made in bacteria. The decreased frameshifting was likely caused by a decreased slippage of the peptidyl-tRNA due to increased availability of aminoacylated tRNA cognate for the A-site codon.
Although none of the above investigations have been pursued sufficiently to establish the detailed molecular mechanism, they collectively demonstrate that frameshifting in these unicellular organisms is sensitive to growth phase. It seems likely that other physiological changes also affect frameshifting. Known physiological changes include oxidative stress, temperature, pH, osmolarity variations, developmental changes (there are large changes in the tRNAs of Bombyx silk glands), irradiation, aging, etc. While so far no studies of possible effects of frameshifting have been done for the great majority of these, a start has been made with certain other changes, as described in the next section.
Elevated incidental frameshifting and its consequences.
Shift-prone sequences not involved in programmed frameshifting are generally not avoided except in highly expressed genes (118). Frameshifting at least at a subset of such sites, and other nonstandard events, can be elevated by perturbations of the balance of tRNAs, when an mRNA with an especially rare codon is highly overexpressed, when polyamine levels become aberrant (18, 19, 133), and with aberrant 2′-5′ oligoadenylates, which lead to altered interferon levels (182). Other causative effects are viral infection (30), cancer, certain triplet repeat expansions (312, 343), and defective ribosomes, whose presence may be more likely in aging cells. The resulting trans-frame products in higher organisms may not previously have been seen by the immune system, and their peptides may be displayed by CD8+ T-cell antigen (278, 362) to give a protective cytotoxic T lymphocyte response or, in the extreme, autoimmunity.
NEW CODONS FOR CODON EXPANSION
The dawn of protein synthesis likely involved specification of a very few amino acids (see, e.g., reference 313). Speculation about the early codon repertoire is more complex, but it may well have involved an expansion of codon usage (see reference 302 and references therein), with UGA encoding selenocysteine (3, 59, 361). Given that UGA is a termination codon in the standard codon table, it is easy to regard selenocysteine specification as context-dependent dynamic redefinition of the meaning of UGA, even though UGA likely encoded selenocysteine before it acquired a termination meaning (reviewed in reference 9). (UAG specification of pyrrolysine [158, 191] is less defined, and a small number of codons have context-independent reassignment of codon meaning. Examples are in the decoding of certain mitochondrial and ciliate nuclear genomes [165].) However, human intervention to expand the repertoire of directly encoded amino acids has several potential advantages and is being vigorously explored (328). Major challenges, especially in vivo, include delivery of the desired unnatural amino acids to aminoacyl-tRNA synthetases, unique recognition of the desired amino acid by a synthetase, and aminoacylation of a tRNA which specifically decodes a designated codon. Ideally, the designated codon needs to be minimally recognized, or not recognized, by a natural tRNA or release factor. One ambitious approach involves the utilization of a new nucleobase pair (201). Another involves using an organism with an unassigned codon(s) or synthesis of a genome to lack specific codons and tRNAs for naturally encoded amino acids that would decode them. A simpler approach, though with obvious limitations, is to use variants of a UAG or UGA suppressor tRNA. Finally, and relevant here, is the use of quadruplet codons (5-7, 137-139, 194, 210, 211, 215, 243, 305-308). Although several of these authors claimed that the tRNA derivatives utilized a quadruplet anticodon, the results reviewed here make this far from a foregone conclusion.
When what is normally a stop codon is used to specify an unnatural amino acid, either as a triplet codon or as the first three bases of a quadruplet codon, then the efficiency of incorporating the novel amino acid can be improved by using specialized ribosomes. One approach has been to have the mRNA that contains the codons specifying the novel amino acids have a variant Shine-Dalgarno sequence which is recognized specifically by suitable specialized ribosomes. The most efficient to date of these have decreased affinity for release factor such that when a stop codon is in the A site, there is a greater chance than normal of a tRNA being accepted instead of the release factor (327).
FRAMESHIFT MUTANT SUPPRESSORS THAT DO NOT INFLUENCE FRAMESHIFTING
Importance of Yeast UPF Mutants
The suppressors in this category do not involve tRNA “gripping” (but if there is no mRNA, there is nothing for the anticodon to pair with!) but have generated very important findings of “gripping” interest. The star performers are the enhancers, actually allosuppressors, of Saccharomyces cerevisiae frameshift mutant suppressors isolated by Mike Culbertson and his colleagues. Following the initial studies (74, 177, 178), the products of genes encoding these enhancers and their counterparts in other organisms have been intensively studied (reviewed in reference 70). They originated with studies of the suppression of the his4-38 +1 frameshift mutant by a particular mutant tRNAGly (206), which occurs at 30°C but not at 37°C. Incubating the combination at the nonpermissive temperature yielded secondary mutations, termed UPF for “up-frameshift.” In the absence of the suppressor tRNA, UPF mutants lack suppressor activity. Disruption of UPF genes leads to stabilization of mRNAs containing a premature in-frame stop codon, either in the WT frame and so arising from a substitution mutation or in a new frame following the site of a frameshift mutation. The products of WT UPF genes, together with eRF1 and eRF3, function to greatly reduce mRNAs containing frameshift mutations or in-frame premature stop codons and thus to reduce wasteful or harmful translation.
Independent work revealed UPF orthologues in Caenorhabditis elegans (257), where there are seven genes (53, 116, 155). The larger number in C. elegans prompted a search for additional genes in S. cerevisiae in the presence of duplications of UPF1, UPF2, and UPF3 so as to avoid mutations in those masking the identification of others. Cosuppressors of a missense mutation and a frameshift mutation were sought in a strain carrying duplications of UPF1, UPF2, and UPF3. One new category was in the EBS1 gene, which encodes a negative regulator of genes whose expression is controlled by the Upf proteins (97). Prior to this and independently, the rapid decay of mammalian mRNAs with premature stop codons was studied (195), and mammalian UPF genes were identified. The mRNA containing the defect in many individuals with human genetic disease is reduced due to the action of the Upf proteins, which consequently are relevant to several disease amelioration strategies that are under development.
In addition to the role of Upf proteins in mRNA surveillance, they also control the decay rate of more than 200 WT S. cerevisiae mRNAs (102, 126, 181). Counterpart proteins are important in development in Drosophila (207). Arabidopsis (8) and many WT mammalian genes are now known (162).
While Upf proteins are in eukaryotes, transcription and translation are coupled in bacteria, with obvious relevance for mRNA stability. Bacterial ribosomes encountering a premature stop codon terminate and dissociate from the mRNA. Though RNA polymerase continues transcription in the absence of closely following ribosomes, the escape from coupling is only transient, as Rho protein can access the mRNA no longer occluded by ribosomes trailing the polymerase. The speed of Rho progression on mRNA allows it to quickly catch up with the polymerase and mediate transcription termination, with mRNA degradation rapidly ensuing. Thus, inactivation of Rho allows continued mRNA synthesis. Consequently, despite the mechanistic distinctions, rho mutants might be expected to act as counterparts to yeast upf mutants and arise as allosuppressors for tRNA mutants, which mediate frameshift mutant suppression. Indeed, among several suppressors of +1 frameshift mutations in the his operon, some of them had an altered Rho factor. The defective Rho factor most likely decreases the polarity induced by the frameshift mutation in the his operon and thereby increases the level of the his mRNA, making the cell independent of His in the growth medium (J. Näsvall and G. R. Björk, unpublished results).
Restoration of Defective Growth Due to Frameshift Suppressors by Secondary Mutations
Members of this category are not allosuppressors, as they do not enhance frameshifting, but because of parallels to allosuppressors, they fit in this section. As described above, one class of suppressors of the −1 frameshift mutant trpE91 had just a substitution of C74 of one the four E. coli tRNA1Val genes (239). The substitution mutants cause a reduction in cell growth. Secondary mutations that restore rapid cell growth have been isolated. The mapped mutations are in a gene originally known as moc (239) but have since been shown by M. O'Connor (cited in reference 13) to be alleles of a gene independently termed hrpA (212). hrpA encodes a 146-kDa protein with similarities to a DEAH box-containing RNA helicase (168a, 212). The suppressor mutant tRNA1Val is specifically shortened, and almost certainly inactivated, by the moc allele studied but not by its WT counterpart (239). This illustrates a different aspect of the potential of suppressor modifiers to reveal unexpected processing pathways.
CONCLUDING REMARKS: READING FRAME MAINTENANCE AND TRANSLOCATION, TWO DIFFERENT FEATURES OF THE RIBOSOME
This review summarizes old and new data concerning how the ribosome maintains the reading frame. The results discussed have been obtained using different biological systems, such as bacteria, yeast, and multicellular organisms. Since translation, including how the ribosome maintains the reading frame, is thought to have evolved before the three domains of life emerged (349), translation occurs in a similar way in all organisms, and it is relevant to compare and discuss data obtained from various biological systems. It is a daunting task for the translation apparatus to hold the ribosome in the correct reading frame for hundreds to thousands of codons. Failure to do so leads to an erroneous peptide sequence being produced, and usually a stop codon is soon encountered in the new frame. Thus, unlike missense errors, which in many cases are not deleterious for the activity or stability of a protein, all frameshift errors are deleterious, providing a rationale for why they are kept at a lower level than missense errors. Although we know from recent development many details of the mechanism of translation, how the ribosome maintains the reading frame and its relation to translocation are still not known. The data discussed in this review therefore have broad and general biological implications.
Evolutionary questions partially motivated the search for frameshift mutant suppressors as tools to probe whether the translation apparatus was indeed resistant to alterations that caused frameshifting. The hope was that if frameshifting could be simply made to happen, it might also occur naturally and be revealing about a fundamental aspect of decoding and the mechanism of translation. Now it is clear that framing malleability is real, and sophisticated programmed frameshifting is used in gene expression. The functionality of some tRNA mutants that emerged from the suppressor studies, and perhaps especially the stacking inferences, together with the finding from programmed frameshifting of anti-Shine-Dalgarno scanning during the elongation phase of protein synthesis may be pertinent to evolution of the code. Inferences from the study of frameshifting are likely to increase with further understanding of ribosome functioning.
The finding of tRNAs with nonstandard anticodon loop sizes and the experimental evidence reviewed concerning the ribosomal E site are likely relevant to considerations of early evolution of decoding. Also reviewed is genetic evidence for intraribosomal anticodon switching in some tRNAs with expanded anticodon loops, noncognate decoding by specific standard tRNAs, and effects of perturbations of tRNA balance.
A different type of motivation, especially for work on the classical expanded-anticodon-loop tRNAs that suppressed +1 frameshift mutants, was the hope that the study of them would increase understanding of the detailed mechanism of translocation. The yardstick model was one outcome of such thoughts, suggesting that the frameshifting induced by these suppressor tRNAs was due to initial four-base translocation. It was assumed that frameshifting and translocation were different manifestations of the same process. However, the summary presented above of how various alterations of tRNA, rRNA, or ribosomal proteins induce frameshifting has revealed quite another view. Frameshifting is predominately a manifestation of altered P-site realignment, and the studies of suppressors have revealed little or no new knowledge about the mechanism of translocation (apart from skepticism about the yardstick model). Thus, reading frame maintenance and translocation are not two sides of the same “coin” but rather two distinct ribosomal features.
Acknowledgments
This work was supported by grants from Science Foundation Ireland, the National Institutes of Health (GM079523), the Swedish Science Research Council (project BU-29309), and the Carl Trygger Foundation.
We are grateful to Phil Farabaugh, Baltimore, MD, and to Michael O'Connor, Kansas City, MO, for their constructive criticisms of the manuscript; to Joakim Näsvall, Umeå, for his help with several figures; to Shahla Thompson; and to our laboratory colleagues present and former, who made our share of this work practical and enjoyable. Figure 1 and the essence of Fig. 3 were kindly provided by Maria Selmer, Uppsala, and Jamie Cate, Berkeley, respectively.
We dedicate this review to the memory of Manny (E. J.) Murgola, who died at its page proof stage.
REFERENCES
- 1.Adamski, F. M., J. F. Atkins, and R. F. Gesteland. 1996. Ribosomal protein L9 interactions with 23 S rRNA: the use of a translational bypass assay to study the effect of amino acid substitutions. J. Mol. Biol. 261357-371. [DOI] [PubMed] [Google Scholar]
- 2.Adamski, F. M., B. C. Donly, and W. P. Tate. 1993. Competition between frameshifting, termination and suppression at the frameshift site in the Escherichia coli release factor-2 mRNA. Nucleic Acids Res. 215074-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambrogelly, A., S. Palioura, and D. Söll. 2007. Natural expansion of the genetic code. Nat. Chem. Biol. 329-35. [DOI] [PubMed] [Google Scholar]
- 4.Ames, B. N., and H. J. Whitfield. 1966. Frameshift mutagenesis in Salmonella. Cold Spring Harb. Symp. Quant. Biol. 31221-225. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, J. C., T. J. Magliery, and P. G. Schultz. 2002. Exploring the limits of codon and anticodon size. Chem. Biol. 9237-244. [DOI] [PubMed] [Google Scholar]
- 6.Anderson, J. C., and P. G. Schultz. 2003. Adaptation of an orthogonal archaeal leucyl-tRNA and synthetase pair for four-base, amber, and opal suppression. Biochemistry 429598-9608. [DOI] [PubMed] [Google Scholar]
- 7.Anderson, J. C., N. Wu, S. W. Santoro, V. Lakshman, D. S. King, and P. G. Schultz. 2004. An expanded genetic code with a functional quadruplet codon. Proc. Natl. Acad. Sci. USA 1017566-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arciga-Reyes, L., L. Wootton, M. Kieffer, and B. Davies. 2006. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47480-489. [DOI] [PubMed] [Google Scholar]
- 9.Atkins, J. F., A. Böck, S. Matsufuji, and R. F. Gesteland. 1999. Dynamics of the genetic code, p. 637-673. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 10.Atkins, J. F., D. Elseviers, and L. Gorini. 1972. Low activity of β-galactosidase in frameshift mutants of Escherichia coli. Proc. Natl. Acad. Sci. USA 691192-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atkins, J. F., and R. F. Gesteland. 1995. Discontinuous triplet decoding with or without re-pairing by peptidyl tRNA, p. 471-490. In D. Söll and U. RajBhandary (ed.), tRNA: structure, biosynthesis, and function. ASM Press, Washington, DC.
- 12.Atkins, J. F., R. F. Gesteland, B. R. Reid, and C. W. Anderson. 1979. Normal tRNAs promote ribosomal frameshifting. Cell 181119-1131. [DOI] [PubMed] [Google Scholar]
- 13.Atkins, J. F., A. J. Herr, C. Massire, M. O'Connor, I. Ivanov, and R. F. Gesteland. 2000. Poking a hole in the sanctity of the triplet code: inferences for framing, p. 369-383. In R. A. Garrett, S. R. Douthwaite, A. Liljas, A. T. Matheson, P. B. Moore, and H. F. Noller (ed.), The ribosome: structure, function, and cellular interaction. American Society for Microbiology, Washington, DC.
- 14.Atkins, J. F., B. P. Nichols, and S. Thompson. 1983. The nucleotide sequence of the first externally suppressible −1 frameshift mutant, and of some nearby leaky frameshift mutants. EMBO J. 21345-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atkins, J. F., and S. Ryce. 1974. UGA and non-triplet suppressor reading of the genetic code. Nature 249527-530. [DOI] [PubMed] [Google Scholar]
- 16.Atkins, J. F., R. B. Weiss, S. Thompson, and R. F. Gesteland. 1991. Towards a genetic dissection of the basis of triplet decoding, and its natural subversion: programmed reading frame shifts and hops. Annu. Rev. Genet. 25201-228. [DOI] [PubMed] [Google Scholar]
- 17.Atkins, J. F., P. V. Baranov, O. Fayet, A. J. Herr, M. T. Howard, I. P. Ivanov, S. Matsufuji, W. A. Miller, B. Moore, M. F. Prere, N. M. Wills, J. Zhou, and R. F. Gesteland. 2001. Overriding standard decoding: implications of recoding for ribosome function and enrichment of gene expression. Cold Spring Harbor Symp. Quant. Biol. 66217-232. [DOI] [PubMed] [Google Scholar]
- 18.Balasundaram, D., J. D. Dinman, C. W. Tabor, and H. Tabor. 1994. SPE1 and SPE2: two essential genes in the biosynthesis of polyamines that modulate +1 ribosomal frameshifting in Saccharomyces cerevisiae. J. Bacteriol. 1767126-7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balasundaram, D., J. D. Dinman, R. B. Wickner, C. W. Tabor, and H. Tabor. 1994. Spermidine deficiency increases +1 ribosomal frameshifting efficiency and inhibits Ty1 retrotransposition in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 91172-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barak, Z., J. Gallant, D. Lindsley, B. Kwieciszewki, and D. Heidel. 1996. Enhanced ribosome frameshifting in stationary phase cells. J. Mol. Biol. 263140-148. [DOI] [PubMed] [Google Scholar]
- 21.Barak, Z., D. Lindsley, and J. Gallant. 1996. On the mechanism of leftward frameshifting at several hungry codons. J. Mol. Biol. 256676-684. [DOI] [PubMed] [Google Scholar]
- 22.Baranov, P. V., R. F. Gesteland, and J. F. Atkins. 2002. Release factor 2 frameshifting sites in different bacteria. EMBO Rep. 3373-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baranov, P. V., R. F. Gesteland, and J. F. Atkins. 2002. Recoding: translational bifurcations in gene expression. Gene 286187-201. [DOI] [PubMed] [Google Scholar]
- 24.Baranov, P. V., R. F. Gesteland, and J. F. Atkins. 2004. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA 10221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnett, L., S. Brenner, F. H. C. Crick, R. G. Shulman, and R. J. Watts-Tobin. 1967. Phase-shift and other mutants in the 1st part of the rIIB cistron of bacteriophage T4. Philos. Trans. R. Soc. London B 252487. [Google Scholar]
- 26.Barrell, B. G., A. R. Coulson, and W. H. McClain. 1973. Nucleotide sequence of a glycine transfer RNA coded by bacteriophage T4. FEBS Lett. 3764-69. [DOI] [PubMed] [Google Scholar]
- 27.Bauerle, R. H., and P. Margolin. 1966. A multifunctional enzyme complex in the tryptophan pathway of Salmonella typhimurium: comparison of polarity and pseudopolarity mutations. Cold Spring Harbor Symp. Quant. Biol. 31203-214. [DOI] [PubMed] [Google Scholar]
- 28.Belcourt, M. F., and P. J. Farabaugh. 1990. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell 62339-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg, O. G., and C. G. Kurland. 1997. Growth rate-optimised tRNA abundance and codon usage. J. Mol. Biol. 270544-550. [DOI] [PubMed] [Google Scholar]
- 30.Berglund, P., D. Finzi, J. R. Bennink, and J. W. Yewdell. 2007. Viral alteration of cellular translational machinery increases defective ribosomal products. J. Virol. 817220-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beringer, M., and M. V. Rodnina. 2007. The ribosomal peptidyl transferase. Mol. Cell 26311-321. [DOI] [PubMed] [Google Scholar]
- 32.Berk, V., and J. H. Cate. 2007. Insights into protein biosynthesis from structures of bacterial ribosomes. Curr. Opin. Struct. Biol. 17302-309. [DOI] [PubMed] [Google Scholar]
- 33.Berk, V., W. Zhang, R. D. Pai, and J. H. Doudna Cate. 2006. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl. Acad. Sci. USA 10315830-15834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berkowitz, D., J. M. Hushon, H. J. Whitfield, J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Besecker, M. I., C. L. Furness, D. M. Coen, and A. Griffiths. 2007. Expression of extremely low levels of thymidine kinase from an acyclovir-resistant herpes simplex virus mutant supports reactivation from latently infected mouse trigeminal ganglia. J. Virol. 818356-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Björk, G. R. 1995. Biosynthesis and function of modified nucleosides in tRNA, p. 165-205. In D. Söll and U. L. RajBhandary (ed.), tRNA: structure, biosynthesis, and function. ASM Press, Washington, DC.
- 37.Björk, G. R., and T. G. Hagervall. 2005. 25 July 2005, posting date. Chapter 4.6.2, Transfer RNA modification. In A. Böck, R. Curtiss III, J. B. Kaper, F. C. Neidhardt, T. Nyström, and C. L. Squires (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington DC. http://www.ecosal.org.
- 38.Björk, G. R., B. Huang, O. P. Persson, and A. S. Byström. 2007. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 131245-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Björk, G. R., K. Jacobsson, K. Nilsson, M. J. O. Johansson, A. S. Byström, and O. P. Persson. 2001. A primordial tRNA modification required for the evolution of life? EMBO J. 20231-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Björk, G. R., P. M. Wikström, and A. S. Byström. 1989. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 244986-989. [DOI] [PubMed] [Google Scholar]
- 41.Blaha, G., and K. H. Nierhaus. 2001. Features and functions of the ribosomal E site. Cold Spring Harbor Symp. Quant. Biol. 66135-146. [DOI] [PubMed] [Google Scholar]
- 42.Blinkova, A., C. Hervas, P. T. Stukenberg, R. Onrust, M. E. O'Donnell, and J. R. Walker. 1993. The Escherichia coli DNA polymerase III holoenzyme contains both products of the dnaX gene, tau and gamma, but only tau is essential. J. Bacteriol. 1756018-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonekamp, F., H. Dalböge, T. Christensen, and K. F. Jensen. 1989. Translation rates of individual codons are not correlated with tRNA abundances or with frequencies of utilization in Escherichia coli. J. Bacteriol. 1715812-5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bossi, L., T. Kohno, and J. R. Roth. 1983. Genetic characterization of the sufJ frameshift suppressor in Salmonella typhimurium. Genetics 10331-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bossi, L., and J. R. Roth. 1981. Four-base codons ACCA, ACCU and ACCC are recognized by frameshift uppressor sufJ. Cell 25489-496. [DOI] [PubMed] [Google Scholar]
- 46.Bossi, L., and D. M. Smith. 1984. Suppressor sufJ: a novel type of tRNA mutant that induces translational frameshifting. Proc. Natl. Acad. Sci. USA 816105-6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bregeon, D., V. Colot, M. Radman, and F. Taddei. 2001. Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev. 152295-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brierley, I., M. R. Meredith, A. J. Bloys, and T. G. Hagervall. 1997. Expression of a coronavirus ribosomal frameshift signal in Escherichia coli: influence of tRNA anticodon modification on frameshifting. J. Mol. Biol. 270360-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bruce, A. G., J. F. Atkins, and R. F. Gesteland. 1986. tRNA anticodon replacement experiments show that ribosomal frameshifting can be caused by doublet decoding. Proc. Natl. Acad. Sci. USA 835062-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brunelle, J. L., E. M. Youngman, D. Sharma, and R. Green. 2006. The interaction between C75 of tRNA and the A loop of the ribosome stimulates peptidyl transferase activity. RNA 1233-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bucklin, D. J., N. M. Wills, R. F. Gesteland, and J. F. Atkins. 2005. P-site pairing subtleties revealed by the effects of different tRNAs on programmed translational bypassing where anticodon re-pairing to mRNA is separated from dissociation. J. Mol. Biol. 34539-49. [DOI] [PubMed] [Google Scholar]
- 52.Calderone, T. L., R. D. Stevens, and T. G. Oas. 1996. High-level misincorporation of lysine for arginine at AGA codons in a fusion protein expressed in Escherichia coli. J. Mol. Biol. 262407-412. [DOI] [PubMed] [Google Scholar]
- 53.Cali, B. M., S. L. Kuchma, J. Latham, and P. Anderson. 1999. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics 151605-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carlson, B. A., S. Y. Kwon, B. J. Lee, and D. Hatfield. 2000. Yeast asparagine (Asn) tRNA without Q base promotes eukaryotic frameshifting more efficiently than mammalian Asn tRNAs with or without Q base. Mol. Cells 10113-118. [DOI] [PubMed] [Google Scholar]
- 55.Carlson, B. A., B. J. Lee, and D. L. Hatfield. 2008. Ribosomal frameshifting in response to hypomodified tRNAs in Xenopus oocytes. Biochem. Biophys. Res. Commun. 37586-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carlson, B. A., J. F. Mushinski, D. W. Henderson, S. Y. Kwon, P. F. Crain, B. J. Lee, and D. L. Hatfield. 2001. 1-Methylguanosine in place of Y base at position 37 in phenylalanine tRNA is responsible for its shiftiness in retroviral ribosomal frameshifting. Virology 279130-135. [DOI] [PubMed] [Google Scholar]
- 57.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. MorganWarren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407340-348. [DOI] [PubMed] [Google Scholar]
- 58.Cashel, M., and K. E. Rudd. 1987. The stringent response, p. 1410-1438. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, DC.
- 59.Chambers, I., J. Frampton, P. Goldfarb, N. Affara, W. McBain, and P. R. Harrison. 1986. The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the ‘termination’ codon, TGA. EMBO J. 51221-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen, P., P. F. Crain, S. J. Näsvall, S. C. Pomerantz, and G. R. Björk. 2005. A “gain of function” mutation in a protein mediates production of novel modified nucleosides. EMBO J. 241842-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Claesson, C., F. Lustig, T. Borén, C. Simonsson, M. Barciszewska, and U. Lagerkvist. 1995. Glycine codon discrimination and the nucleotide in position 32 of the anticodon loop. J. Mol. Biol. 247191-196. [DOI] [PubMed] [Google Scholar]
- 62.Clare, J., and P. Farabaugh. 1985. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc. Natl. Acad. Sci. USA 822829-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62a.Cornish, P. V., D. N. Ermolenko, D. W. Staple, R. P. Hickerson, H. F. Noller, and T. Ha. 3 February 2009, posting date. Following movement of the L1 stalk between three functional states in single ribosomes. Proc. Natl. Acad. Sci. USA. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed]
- 63.Cortazzo, P., C. Cerveñansky, M. Marín, C. Reiss, R. Ehrlich, and A. Deana. 2002. Silent mutations affect in vivo protein folding in Escherichia coli. Biochem. Biophys. Res. Commun. 293537-541. [DOI] [PubMed] [Google Scholar]
- 64.Craigen, W. J., and C. T. Caskey. 1986. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature 322273-275. [DOI] [PubMed] [Google Scholar]
- 65.Crick, F. H., S. Brenner, A. Klug, and G. Pieczenik. 1976. A speculation on the origin of protein synthesis. Origins Life 7389-397. [DOI] [PubMed] [Google Scholar]
- 66.Crick, F. H. C. 1957. The structure of nucleic acids and their role in protein synthesis. Biochem. Soc. Symp. 1425-26. [PubMed] [Google Scholar]
- 67.Crick, F. H. C. 1988. What mad pursuit. A personal view of scientific discovery, chapter 12. Triplets. Alfred P. Sloan Foundation Series, Basic Books Inc., Publishers. New York, NY.
- 68.Crick, F. H. C., L. Barnett, S. Brenner, and R. J. Watts-Tobin. 1961. General nature of the genetic code for proteins. Nature 1921227-1232. [DOI] [PubMed] [Google Scholar]
- 69.Cukras, A. R., D. R. Southworth, J. L. Brunelle, G. M. Culver, and R. Green. 2003. Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol. Cell 12321-328. [DOI] [PubMed] [Google Scholar]
- 70.Culbertson, M. R. 2007. Navigating without a road map. Genetics 1771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Culbertson, M. R., L. Charnas, M. T. Johnson, and G. R. Fink. 1977. Frameshifts and frameshift suppressors in Saccharomyces cerevisiae. Genetics 86745-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Culbertson, M. R., R. F. Gaber, and C. M. Cummins. 1982. Frameshift suppression in Saccharomyces cerevisiae. V. Isolation and genetic properties of nongroup-specific suppressors. Genetics 102361-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Culbertson, M. R., P. Leeds, M. G. Sandbaken, and P. G. Wilson. 1990. Frameshift suppression, p. 527-533. In W. E. Hill, A. Dahlberg, R. A. Garrett, P. B. Moore, D. Schlessinger, and J. R. Warner (ed.), The ribosome: structure, function, and evolution. American Society for Microbiology, Washington, DC.
- 74.Culbertson, M. R., K. M. Underbrink, and G. R. Fink. 1980. Frameshift suppression Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics 95833-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cummins, C. M., M. R. Culbertson, and G. Knapp. 1985. Frameshift suppressor mutations outside the anticodon in yeast proline tRNAs containing an intervening sequence. Mol. Cell. Biol. 51760-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cummins, C. M., T. F. Donahue, and M. R. Culbertson. 1982. Nucleotide sequence of the SUF2 frameshift suppressor gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 793565-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cummins, C. M., R. F. Gaber, M. R. Culbertson, R. Mann, and G. R. Fink. 1980. Frameshift suppression in Saccharomyces cerevisiae. III. Isolation and genetic properties of group III suppressors. Genetics 95855-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Curran, J. F. 1995. Decoding with the A-I wobble pair is inefficient. Nucleic Acids Res. 23683-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Curran, J. F., and M. Yarus. 1986. Base substitutions in the tRNA anticodon arm do not degrade the accuracy of reading frame maintenance. Proc. Natl. Acad. Sci. USA 836538-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Czworkowski, J., and P. B. Moore. 1997. The conformational properties of elongation factor G and the mechanism of translocation. Biochemistry 3610327-10334. [DOI] [PubMed] [Google Scholar]
- 81.Dayhuff, T. J., J. F. Atkins, and R. F. Gesteland. 1986. Characterization of ribosomal frameshift events by protein sequence analysis. J. Biol. Chem. 2617491-7500. [PubMed] [Google Scholar]
- 82.DeLano, W. L. 2002. The PyMOL molcecular graphics system. DeLano Scientific, Palo Alto, CA.
- 83.Di Giulio, M. 2008. Why the genetic code originated. Implications of the origin of protein synthesis, p. 59-67. In M. Barbieri (ed.), The codes of life: the rules of macroevolution. Springer, New York, NY.
- 84.Dinman, J. D., and T. G. Kinzy. 1997. Translational misreading: mutations in translation elongation factor 1α differentially affect programmed ribosomal frameshifting and drug sensitivity. RNA 3870-881. [PMC free article] [PubMed] [Google Scholar]
- 85.Doel, S. M., S. J. McCready, C. R. Nierras, and B. S. Cox. 1994. The dominant PNM2- mutation which eliminates the psi factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics 137659-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Donahue, T. F., P. J. Farabaugh, and G. R. Fink. 1981. Suppressible four-base glycine and proline codons in yeast. Science 212455-457. [DOI] [PubMed] [Google Scholar]
- 87.Dong, H. J., L. Nilsson, and C. G. Kurland. 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260649-663. [DOI] [PubMed] [Google Scholar]
- 88.Dunham, C. M., M. Selmer, S. S. Phelps, A. C. Kelley, T. Suzuki, S. Joseph, and V. Ramakrishnan. 2007. Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit. RNA 13817-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dunn, J. J., and F. W. Studier. 1983. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 166477-535. [DOI] [PubMed] [Google Scholar]
- 90.Erbe, R. W., M. M. Nau, and P. Leder. 1969. Translation and translocation of defined RNA messengers. J. Mol. Biol. 39441-460. [DOI] [PubMed] [Google Scholar]
- 91.Fahlman, R. P., T. Dale, and O. C. Uhlenbeck. 2004. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell 16799-805. [DOI] [PubMed] [Google Scholar]
- 92.Farabaugh, P. J. 1996. Programmed translational frameshifting. Microbiol. Rev. 60103-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Farabaugh, P. J. 1997. Programmed alternative reading of the genetic code. R. G. Landes Company, Austin, TX.
- 94.Farabaugh, P. J., and G. R. Björk. 1999. How translational accuracy influences reading frame maintenance. EMBO J. 181427-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Farabaugh, P. J., and A. Vimaladithan. 1998. Effect of frameshift-inducing mutants of elongation factor 1 alpha on programmed +1 frameshifting in yeast. RNA 438-46. [PMC free article] [PubMed] [Google Scholar]
- 96.Farabaugh, P. J., H. Zhao, and A. Vimaladithan. 1993. A novel programed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell 7493-103. (Erratum, 75:826.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ford, A. S., Q. Guan, E. Neeno-Eckwall, and M. R. Culbertson. 2006. Ebs1p, a negative regulator of gene expression controlled by the Upf proteins in the yeast Saccharomyces cerevisiae. Eukaryot. Cell 5301-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fox, T. D., and B. Weiss-Brummer. 1980. Leaky +1 and −1 frameshift mutations at the same site in a yeast mitochondrial gene. Nature 28860-63. [DOI] [PubMed] [Google Scholar]
- 99.Fredrick, K., and H. F. Noller. 2003. Catalysis of ribosomal translocation by sparsomycin. Science 3001159-1162. [DOI] [PubMed] [Google Scholar]
- 100.Fu, C., and J. Parker. 1994. A ribosomal frameshifting error during translation of the argI mRNA of Escherichia coli. Mol. Gen. Genet. 243434-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fuller, W., and A. Hodgson. 1967. Conformation of the anticodon loop intRNA. Nature 215817-821. [DOI] [PubMed] [Google Scholar]
- 102.Gaba, A., A. Jacobson, and M. S. Sachs. 2005. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell 20449-460. [DOI] [PubMed] [Google Scholar]
- 103.Gaber, R. F., and M. R. Culbertson. 1982. The yeast frameshift suppressor gene SUF16-1 encodes an altered glycine tRNA containing the four-base anticodon 3′-CCCG-5′. Gene 19163-172. [DOI] [PubMed] [Google Scholar]
- 104.Gaber, R. F., and M. R. Culbertson. 1984. Codon recognition during frameshift suppression in Saccharomyces cerevisiae. Mol. Cell. Biol. 42052-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gallant, J., P. Bonthuis, D. Lindsley, J. Cabellon, G. Gill, K. Heaton, B. Kelley-Clarke, L. MacDonald, S. Mercer, H. Vu, and A. Worsley. 2004. On the role of the starved codon and the takeoff site in ribosome bypassing in Escherichia coli. J. Mol. Biol. 342713-724. [DOI] [PubMed] [Google Scholar]
- 106.Gallant, J., and D. Foley. 1980. On the causes and prevention of mistranslation, p. 615-638. In G. Chambliss, G. R. Craven, J. Davies, K. Davis, L. Kahan, and M. Nomura (ed.), Ribosome: structure, function and genetics. University Park Press, Baltimore, MD.
- 107.Gallant, J., D. Lindsley, and J. Masucci. 2000. The unbearable lightness of peptidyl-tRNA, p. 385-396. In R. A. Garrett, S. R. Douthwaite, A. Liljas, A. T. Matheson, P. B. Moore, and H. F. Noller (ed.), The ribosome: structure, function, antibiotics, and cellular interaction. American Society for Microbiology, Washington, DC.
- 108.Gavrilova, L. P., O. E. Kostiashkina, V. E. Koteliansky, N. M. Rutkevitch, and A. S. Spirin. 1976. Factor-free (“non-enzymic”) and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J. Mol. Biol. 101537-552. [DOI] [PubMed] [Google Scholar]
- 109.Gavrilova, L. P., and A. S. Spirin. 1971. Stimulation of “non-enzymic” translocation in ribosomes by p-chloromercuribenzoate. FEBS Lett. 17324-326. [DOI] [PubMed] [Google Scholar]
- 110.Gesteland, R. F., and J. F. Atkins. 1996. Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 65741-768. [DOI] [PubMed] [Google Scholar]
- 111.Gesteland, R. F., R. B. Weiss, and J. F. Atkins. 1992. Recoding: reprogrammed genetic decoding. Science 2571640-1641. [DOI] [PubMed] [Google Scholar]
- 112.Gramstat, A., D. Prüfer, and W. Rohde. 1994. The nucleic acid-binding zinc finger protein of potato virus M is translated by internal initiation as well as by ribosomal frameshifting involving a shifty stop codon and a novel mechanism of P-site slippage. Nucleic Acids Res. 223911-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Green, R., and H. F. Noller. 1997. Ribosomes and translation. Annu. Rev. Biochem. 66679-716. [DOI] [PubMed] [Google Scholar]
- 114.Gregory, S. T., K. R. Lieberman, and A. E. Dahlberg. 1994. Mutations in the peptidyl transferase region of E. coli 23S rRNA affecting translational accuracy. Nucleic Acids Res. 22279-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Griffiths, A., S. H. Chen, B. C. Horsburgh, and D. M. Coen. 2003. Translational compensation of a frameshift mutation affecting herpes simplex virus thymidine kinase is sufficient to permit reactivation from latency. J. Virol. 774703-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Grimson, A., S. O'Connor, C. L. Newman, and P. Anderson. 2004. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA Decay in Caenorhabditis elegans. Mol. Cell. Biol. 247483-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gurvich, O. L., P. V. Baranov, R. F. Gesteland, and J. F. Atkins. 2005. Expression levels influence ribosomal frameshifting at the tandem rare arginine codons AGG_AGG and AGA_AGA in Escherichia coli. J. Bacteriol. 1874023-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gurvich, O. L., P. V. Baranov, J. Zhou, A. W. Hammer, R. F. Gesteland, and J. F. Atkins. 2003. Sequences that direct significant levels of frameshifting are frequent in coding regions of Escherichia coli. EMBO J. 225941-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hagervall, T. G., S. C. Pomerantz, and J. A. McCloskey. 1998. Reduced misreading of asparagine codons by Escherichia coli tRNA(Lys) with hypomodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J. Mol. Biol. 28433-42. [DOI] [PubMed] [Google Scholar]
- 120.Hagervall, T. G., T. M. Tuohy, J. F. Atkins, and G. R. Björk. 1993. Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation. J. Mol. Biol. 232756-765. [DOI] [PubMed] [Google Scholar]
- 121.Hansen, J. L., T. M. Schmeing, P. B. Moore, and T. A. Steitz. 2002. Structural insights into peptide bond formation. Proc. Natl. Acad. Sci. USA 9911670-11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hansen, T. M., P. V. Baranov, I. P. Ivanov, R. F. Gesteland, and J. F. Atkins. 2003. Maintenance of the correct open reading frame by the ribosome. EMBO Rep. 4499-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hardesty, B., W. Clup, and W. McKeehan. 1969. The sequence of reactions leading to the synthesis of a peptide bond on reticulocyte ribosome. Cold Spring Harbor Symp. Quant. Biol. 34331-345. [DOI] [PubMed] [Google Scholar]
- 124.Hatfield, D., Y. X. Feng, B. J. Lee, A. Rein, J. G. Levin, and S. Oroszlan. 1989. Chromatographic analysis of the aminoacyl-tRNAs which are required for translation of codons at and around the ribosomal frameshift sites of HIV, HTLV-1, and BLV. Virology 173736-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hatfield, D. L., D. W. Smith, B. J. Lee, P. J. Worland, and S. Oroszlan. 1990. Structure and function of suppressor tRNAs in higher eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2571-96. [DOI] [PubMed] [Google Scholar]
- 126.He, F., X. Li, P. Spatrick, R. Casillo, S. Dong, and A. Jacobson. 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 121439-1452. [DOI] [PubMed] [Google Scholar]
- 127.Hendrick, J. L., P. G. Wilson, I. I. Edelman, M. G. Sandbaken, D. Ursic, and M. R. Culbertson. 2001. Yeast frameshift suppressor mutations in the genes coding for transcription factor Mbf1p and ribosomal protein S3: evidence for autoregulation of S3 synthesis. Genetics 1571141-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Herbst, K. L., L. M. Nichols, R. F. Gesteland, and R. B. Weiss. 1994. A mutation in ribosomal protein L9 affects ribosomal hopping during translation of gene 60 from bacteriophage T4. Proc. Natl. Acad. Sci. USA 9112525-12529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Herr, A. J., J. F. Atkins, and R. F. Gesteland. 1999. Mutations which alter the elbow region of tRNA2Gly reduce T4 gene 60 translational bypassing efficiency. EMBO J. 182886-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Herr, A. J., R. F. Gesteland, and J. F. Atkins. 2000. One protein from two open reading frames: mechanism of a 50 nt translational bypass. EMBO J. 192671-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Herr, A. J., C. C. Nelson, N. M. Wills, R. F. Gesteland, and J. F. Atkins. 2001. Analysis of the roles of tRNA structure, ribosomal protein L9, and the bacteriophage T4 gene 60 bypassing signals during ribosome slippage on mRNA. J. Mol. Biol. 3091029-1048. [DOI] [PubMed] [Google Scholar]
- 132.Herr, A. J., N. M. Wills, C. C. Nelson, R. F. Gesteland, and J. F. Atkins. 2004. Factors that influence selection of coding resumption sites in translational bypassing: minimal conventional peptidyl-tRNA:mRNA pairing can suffice. J. Biol. Chem. 27911081-11087. [DOI] [PubMed] [Google Scholar]
- 133.Higashi, K., K. Kashiwagi, S. Taniguchi, Y. Terui, K. Yamamoto, A. Ishihama, and K. Igarashi. 2006. Enhancement of +1 frameshift by polyamines during translation of polypeptide release factor 2 in Escherichia coli. J. Biol. Chem. 2819527-9537. [DOI] [PubMed] [Google Scholar]
- 134.Hill, C. W., G. Combriato, W. Steinhart, D. L. Riddle, and J. Carbon. 1973. The nucleotide sequence of the GGG-specific glycine transfer ribonucleic acid of Escherichia coli and of Salmonella typhimurium. J. Biol. Chem. 2484252-4262. [PubMed] [Google Scholar]
- 135.Hizi, A., L. E. Henderson, T. D. Copeland, R. C. Sowder, C. V. Hixson, and S. Oroszlan. 1987. Characterization of mouse mammary tumor virus gag-pro gene products and the ribosomal frameshift site by protein sequencing. Proc. Natl. Acad. Sci. USA 847041-7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hoang, L., K. Fredrick, and H. F. Noller. 2004. Creating ribosomes with an all-RNA 30S subunit P site. Proc. Natl. Acad. Sci. USA 10112439-12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hohsaka, T., Y. Ashizuka, H. Murakami, and M. Sisido. 2001. Five-base codons for incorporation of nonnatural amino acids into proteins. Nucleic Acids Res. 293646-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hohsaka, T., Y. Ashizuka, H. Taira, H. Murakami, and M. Sisido. 2001. Incorporation of nonnatural amino acids into proteins by using various four-base codons in an Escherichia coli in vitro translation system. Biochemistry 4011060-11064. [DOI] [PubMed] [Google Scholar]
- 139.Hohsaka, T., N. Muranaka, C. Komiyama, K. Matsui, S. Takaura, R. Abe, H. Murakami, and M. Sisido. 2004. Position-specific incorporation of dansylated non-natural amino acids into streptavidin by using a four-base codon. FEBS Lett. 560173-177. [DOI] [PubMed] [Google Scholar]
- 140.Holley, R. W., G. A. Everett, J. T. Madison, and A. Zamir. 1965. Nucleotide sequences in the yeast alanine transfer ribonucleic acid. J. Biol. Chem. 2402122-2128. [PubMed] [Google Scholar]
- 141.Houssier, C., P. Degree, K. Nicoghosian, and H. Grosjean. 1988. Effect of uridine dethiolation in the anticodon triplet of tRNA(Glu) on its association with tRNA(Phe). J. Biomol. Struct. Dyn. 51259-1266. [DOI] [PubMed] [Google Scholar]
- 142.Hughes, D., J. F. Atkins, and S. Thompson. 1987. Mutants of elongation factor Tu promote ribosomal frameshifting and nonsense readthrough. EMBO J. 64235-4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hüttenhofer, A., B. Weiss-Brummer, G. Dirheimer, and R. P. Martin. 1990. A novel type of + 1 frameshift suppressor: a base substitution in the anticodon stem of a yeast mitochondrial serine-tRNA causes frameshift suppression. EMBO J. 9551-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ikemura, T. 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 1461-21. [DOI] [PubMed] [Google Scholar]
- 145.Ikemura, T. 1982. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting trans. J. Mol. Biol. 158573-597. [DOI] [PubMed] [Google Scholar]
- 146.Ikemura, T. 1985. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 213-34. [DOI] [PubMed] [Google Scholar]
- 147.Ikemura, T., and H. Ozeki. 1983. Codon usage and transfer RNA contents: organism-specific codon-choice patterns in reference to the isoacceptor contents. Cold Spring Harbor Symp. Quant. Biol. 471087-1097. [DOI] [PubMed] [Google Scholar]
- 148.Inoue-Yokosawa, N., C. Ishikawa, and Y. Kaziro. 1974. The role of guanosine triphosphate in translocation reaction catalyzed by elongation factor G. J. Biol. Chem. 2494321-4323. [PubMed] [Google Scholar]
- 149.Ito, K., M. Uno, and Y. Nakamura. 2000. A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature 403680-684. [DOI] [PubMed] [Google Scholar]
- 150.Jacks, T., H. D. Madhani, F. R. Masiarz, and H. E. Varmus. 1988. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell 55447-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jacks, T., K. Townsley, H. E. Varmus, and J. Majors. 1987. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc. Natl. Acad. Sci. USA 844298-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jacks, T., and H. E. Varmus. 1985. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science 2301237-1242. [DOI] [PubMed] [Google Scholar]
- 153.Jenner, L., B. Rees, M. Yusupov, and G. Yusupova. 2007. Messenger RNA conformations in the ribosomal E site revealed by X-ray crystallography. EMBO Rep. 8846-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Johansson, M. J., A. Esberg, B. Huang, G. R. Björk, and A. S. Byström. 2008. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol. 283301-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Johns, L., A. Grimson, S. L. Kuchma, C. L. Newman, and P. Anderson. 2007. Caenorhabditis elegans SMG-2 selectively marks mRNAs containing premature translation termination codons. Mol. Cell. Biol. 275630-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kanaya, S., Y. Yamada, M. Kinouchi, Y. Kudo, and T. Ikemura. 2001. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J. Mol. Evol. 53290-298. [DOI] [PubMed] [Google Scholar]
- 157.Kane, J. F., B. N. Violand, D. F. Curran, N. R. Staten, K. L. Duffin, and G. Bogosian. 1992. Novel in-frame two codon translational hop during synthesis of bovine placental lactogen in a recombinant strain of Escherichia coli. Nucleic Acids Res. 206707-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kavran, J. M., S. Gundllapalli, P. O'Donoghue, M. Englert, D. Söll, and T. A. Steitz. 2007. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc. Natl. Acad. Sci. USA 10411268-11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kawakami, K., Y. H. J.önsson, G. R. Björk, H. Ikeda, and Y. Nakamura. 1988. Chromosomal location and structure of the operon encoding peptide-chain-release factor 2 of Escherichia coli. Proc. Natl. Acad. Sci. USA 855620-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kharat, A. S., E. Coursange, M. Noirclerc-Savoye, J. Lacoste, and M. Blot. 2006. IS1 transposition is enhanced by translation errors and by bacterial growth at extreme glucose levels. Acta Biochim. Pol. 53729-738. [PubMed] [Google Scholar]
- 161.Kim, D. F., and R. Green. 1999. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol. Cell 4859-864. [DOI] [PubMed] [Google Scholar]
- 162.Kim, Y. K., L. Furic, M. Parisien, F. Major, L. DesGroseillers, and L. E. Maquat. 2007. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 262670-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kimchi-Sarfaty, C., J. M. Oh, I. W. Kim, Z. E. Sauna, A. M. Calcagno, S. V. Ambudkar, and M. M. Gottesman. 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315525-528. [DOI] [PubMed] [Google Scholar]
- 164.Klobutcher, L. A., and P. J. Farabaugh. 2002. Shifty ciliates: frequent programmed translational frameshifting in euplotids. Cell 111763-766. [DOI] [PubMed] [Google Scholar]
- 165.Knight, R. D., S. J. Freeland, and L. F. Landweber. 2001. Rewiring the keyboard: evolvability of the genetic code. Nat. Rev. Genet. 249-58. [DOI] [PubMed] [Google Scholar]
- 166.Kohno, T., L. Bossi, and J. R. Roth. 1983. New suppressors of frameshift mutations in Salmonella typhimurium. Genetics 10323-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kohno, T., and J. R. Roth. 1978. A Salmonella frameshift suppressor that acts at runs of A residues in the messenger RNA. J. Mol. Biol. 12637-52. [DOI] [PubMed] [Google Scholar]
- 168.Komar, A. A., T. Lesnik, and C. Reiss. 1999. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 462387-391. [DOI] [PubMed] [Google Scholar]
- 168a.Koo, J. T., J. Choe, and S. L. Moseley. 2004. HrpA, a DEAH-box RNA helicase, is involved in mRNA processing of a fimbrial operon in Escherichia coli. Mol. Microbiol. 521813-1826. [DOI] [PubMed]
- 169.Korostelev, A., and H. F. Noller. 2007. Analysis of structural dynamics in the ribosome by TLS crystallographic refinement. J. Mol. Biol. 3731058-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Korostelev, A., and H. F. Noller. 2007. The ribosome in focus: new structures bring new insights. Trends Biochem. Sci. 32434-441. [DOI] [PubMed] [Google Scholar]
- 171.Kramer, E. B., and P. J. Farabaugh. 2007. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 1387-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kurland, C. C. 1979. Reading frame errors on ribosomes, p. 97-108. In J. E. Celis J. D. and Smith (ed.), Nosense mutation and tRNA suppressors. Academic Press, London, United Kingdom.
- 173.Lagerkvist, U. 1978. “Two out of three”: an alternative method for codon reading. Proc. Natl. Acad. Sci. USA 751759-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.LaRiviere, F. J., A. D. Wolfson, and O. C. Uhlenbeck. 2001. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 294165-168. [DOI] [PubMed] [Google Scholar]
- 175.Larsen, B., N. M. Wills, C. Nelson, J. F. Atkins, and R. F. Gesteland. 2000. Nonlinearity in genetic decoding: homologous DNA replicase genes use alternatives of transcriptional slippage or translational frameshifting. Proc. Natl. Acad. Sci. USA 971683-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lecointe, F., O. Namy, I. Hatin, G. Simos, J. P. Rousset, and H. Grosjean. 2002. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J. Biol. Chem. 27730445-30453. [DOI] [PubMed] [Google Scholar]
- 177.Leeds, P., S. W. Peltz, A. Jacobson, and M. R. Culbertson. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 52303-2314. [DOI] [PubMed] [Google Scholar]
- 178.Leeds, P., J. M. Wood, B. S. Lee, and M. R. Culbertson. 1992. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 122165-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Leipuviene, R., and G. R. Björk. 2005. A reduced level of charged tRNAmnm5UCUArg triggers the wild-type peptidyl-tRNA to frameshift. RNA 11796-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Leipuviene, R., and G. R. Björk. 2007. Alterations in the two globular domains or in the connecting α-helix of bacterial ribosomal protein L9 induces +1 frameshifts. J. Bacteriol. 1897024-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lelivelt, M. J., and M. R. Culbertson. 1999. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol. Cell. Biol. 196710-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Le Roy, F., T. Salehzada, C. Bisbal, J. P. Dougherty, and S. W. Peltz. 2005. A newly discovered function for RNase L in regulating translation termination. Nat. Struct. Mol. Biol. 12505-512. [DOI] [PubMed] [Google Scholar]
- 183.Lewin, B. 1983. Genes. John Wiley & Sons, Inc, New York, NY.
- 184.Li, J. N., and G. R. Björk. 1999. Structural alterations of the tRNA(m1G37)methyltransferase from Salmonella typhimurium affect tRNA substrate specificity. RNA 5395-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Li, J. N., B. Esberg, J. F. Curran, and G. R. Björk. 1997. Three modified nucleosides present in the anticodon stem and loop influence the in vivo aa-tRNA selection in a tRNA-dependent manner. J. Mol. Biol. 271209-221. [DOI] [PubMed] [Google Scholar]
- 186.Li, M., and A. Tzagoloff. 1979. Assembly of the mitochondrial membrane system: sequences of yeast mitochondrial valine and an unusual threonine tRNA gene. Cell 1847-53. [DOI] [PubMed] [Google Scholar]
- 187.Liao, P. Y., P. Gupta, A. N. Petrov, J. D. Dinman, and K. H. Lee. 2008. A new kinetic model reveals the synergistic effect of E-, P- and A-sites on +1 ribosomal frameshifting. Nucleic Acids Res. 362619-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Licznar, P., N. Mejlhede, M. F. Prère, N. Wills, R. F. Gesteland, J. F. Atkins, and O. Fayet. 2003. Programmed translational −1 frameshifting on hexanucleotide motifs and the wobble properties of tRNAs. EMBO J. 224770-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Limbach, P. A., P. F. Crain, and J. A. McCloskey. 1994. The modified nucleosides of RNA. Nucleic Acids Res. 222183-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lindsley, D., J. Gallant, C. Doneanu, P. Bonthuis, S. Caldwell, and A. Fontelera. 2005. Spontaneous ribosome bypassing in growing cells. J. Mol. Biol. 349261-272. [DOI] [PubMed] [Google Scholar]
- 191.Longstaff, D. G., S. K. Blight, L. Zhang, K. B. Green-Church, and J. A. Krzycki. 2007. In vivo contextual requirements for UAG translation as pyrrolysine. Mol. Microbiol. 63229-241. [DOI] [PubMed] [Google Scholar]
- 192.Lustig, F., T. Borén, C. Claesson, C. Simonsson, M. Barciszewska, and U. Lagerkvist. 1993. The nucleotide in position 32 of the tRNA anticodon loop determines ability of anticodon UCC to discriminate among glycine codons. Proc. Natl. Acad. Sci. USA 903343-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.MacVanin, M., U. Johanson, M. Ehrenberg, and D. Hughes. 2000. Fusidic acid-resistant EF-G perturbs the accumulation of ppGpp. Mol. Microbiol. 3798-107. [DOI] [PubMed] [Google Scholar]
- 194.Magliery, T. J., J. C. Anderson, and P. G. Schultz. 2001. Expanding the genetic code: selection of efficient suppressors of four-base codons and identification of '′shifty” four-base codons with a library approach in Escherichia coli. J. Mol. Biol. 307755-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Maquat, L. E., A. J. Kinniburgh, E. A. Rachmilewitz, and J. Ross. 1981. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell 27543-553. [DOI] [PubMed] [Google Scholar]
- 196.Marczinke, B., T. Hagervall, and I. Brierley. 2000. The Q-base of asparaginyl-tRNA is dispensable for efficient −1 ribosomal frameshifting in eukaryotes. J. Mol. Biol. 295179-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Márquez, V., D. N. Wilson, W. P. Tate, F. Triana-Alonso, and K. H. Nierhaus. 2004. Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell 11845-55. [DOI] [PubMed] [Google Scholar]
- 198.Masucci, J. P., J. Gallant, D. Lindsley, and J. Atkinson. 2002. Influence of the relA gene on ribosome frameshifting. Mol. Genet. Genomics 26881-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mathison, L., and M. R. Culbertson. 1985. Suppressible and nonsuppressible +1 G-C base pair insertions induced by ICR-170 at the his4 locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 52247-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mathison, L., M. Winey, C. Soref, M. R. Culbertson, and G. Knapp. 1989. Mutations in the anticodon stem affect removal of introns from pre-tRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 94220-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Matsuda, S., J. D. Fillo, A. A. Henry, P. Rai, S. J. Wilkens, T. J. Dwyer, B. H. Geierstanger, D. E. Wemmer, P. G. Schultz, G. Spraggon, and F. E. Romesberg. 2007. Efforts toward expansion of the genetic alphabet: structure and replication of unnatural base pairs. J. Am. Chem. Soc. 12910466-10473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Matzke, A. J., A. Barta, and E. Kuechler. 1980. Mechanism of translocation: relative arrangement of tRNA and mRNA on the ribosome. Proc. Natl. Acad. Sci. USA 775110-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mazumdar, S. K., and W. Saenger. 1974. Molecular structure of poly-2-thiouridylic acid, a double helix with non-equivalent polynucleotide chains. J. Mol. Biol. 85213-219. [DOI] [PubMed] [Google Scholar]
- 205.Mellor, J., S. M. Fulton, M. J. Dobson, W. Wilson, S. M. Kingsman, and A. J. Kingsman. 1985. A retrovirus-like strategy for expression of a fusion protein encoded by yeast transposon Ty1. Nature 313243-246. [DOI] [PubMed] [Google Scholar]
- 206.Mendenhall, M. D., P. Leeds, H. Fen, L. Mathison, M. Zwick, C. Sleiziz, and M. R. Culbertson. 1987. Frameshift suppressor mutations affecting the major glycine transfer RNAs of Saccharomyces cerevisiae. J. Mol. Biol. 19441-58. [DOI] [PubMed] [Google Scholar]
- 207.Metzstein, M. M., and M. A. Krasnow. 2006. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet. 2e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Miranda, I., R. Silva, and M. A. Santos. 2006. Evolution of the genetic code in yeasts. Yeast 23203-213. [DOI] [PubMed] [Google Scholar]
- 209.Moazed, D., and H. F. Noller. 1989. Intermediate states in the movement of transfer RNA in the ribosome. Nature 342142-148. [DOI] [PubMed] [Google Scholar]
- 210.Moore, B., C. C. Nelson, B. C. Persson, R. F. Gesteland, and J. F. Atkins. 2000. Decoding of tandem quadruplets by adjacent tRNAs with eight-base anticodon loops. Nucleic Acids Res. 283615-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Moore, B., B. C. Persson, C. C. Nelson, R. F. Gesteland, and J. F. Atkins. 2000. Quadruplet codons: implications for code expansion and the specification of translation step size. J. Mol. Biol. 298195-209. [DOI] [PubMed] [Google Scholar]
- 212.Moriya, H., H. Kasai, and K. Isono. 1995. Cloning and characterization of the hrpA gene in the terC region of Escherichia coli that is highly similar to the DEAH family RNA helicase genes of Saccharomyces cerevisiae. Nucleic Acids Res. 23595-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Motorin, Y., and H. Grosjean. 1998. Appendix 1: chemical structures and classification of posttranscriptionally modified nucleosides in RNA, p. 543-549. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, DC.
- 214.Munro, J. B., R. B. Altman, N. O'Connor, and S. C. Blanchard. 2007. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell 25505-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Muranaka, N., T. Hohsaka, and M. Sisido. 2006. Four-base codon mediated mRNA display to construct peptide libraries that contain multiple nonnatural amino acids. Nucleic Acids Res. 34e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Murgola, E. J. 1985. tRNA, suppression, and the code. Annu. Rev. Genet. 1957-80. [DOI] [PubMed] [Google Scholar]
- 217.Murgola, E. J., N. E. Prather, B. H. Mims, F. T. Pagel, and K. A. Hijazi. 1983. Anticodon shift in tRNA: a novel mechanism in missense and nonsense suppression. Proc. Natl. Acad. Sci. USA 804936-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Namy, O., A. Galopier, C. Martini, S. Matsufuji, C. Fabret, and J. P. Rousset. 2008. Epigenetic control of polyamines by the prion [PSI(+)]. Nat. Cell Biol. 101069-1075. [DOI] [PubMed] [Google Scholar]
- 219.Namy, O., J.-P. Rousset, S. Napthine, and I. Brierley. 2004. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell 13157-168. [DOI] [PubMed] [Google Scholar]
- 220.Näsvall, S. J., P. Chen, and G. R. Björk. 2004. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAProcmo5UGG promotes reading of all four proline codons in vivo. RNA 101662-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Näsvall, S. J., K. Nilsson, and G. R. Björk. 2009. The ribosomal grip of the peptidyl-tRNA is critical for reading frame maintenance. J. Mol. Biol. 385350-367. [DOI] [PubMed] [Google Scholar]
- 222.Newmark, R. A., and C. R. Cantor. 1968. Nuclear magnetic resonance study of the interactions of guanosine and cytidine in dimethyl sulfoxide. J. Am. Chem. Soc. 905010-5017. [DOI] [PubMed] [Google Scholar]
- 223.Newton, A. 1970. Isolation and characterization of frameshift mutations in the lac operon. J. Mol. Biol. 49589-601. [DOI] [PubMed] [Google Scholar]
- 224.Nissen, P., J. Hansen, N. Ban, P. B. Moore, and T. A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289920-930. [DOI] [PubMed] [Google Scholar]
- 225.Noller, H. F. 2006. Evolution of ribosomes and translation from an RNA world, p. 287-307. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 226.Noller, H. F., M. M. Yusupov, G. Z. Yusupova, A. Baucom, and J. H. Cate. 2002. Translocation of tRNA during protein synthesis. FEBS Lett. 51411-16. [DOI] [PubMed] [Google Scholar]
- 227.O′Connor, M. 1998. tRNA imbalance promotes −1 frameshifting via near-cognate decoding. J. Mol. Biol. 279727-736. [DOI] [PubMed] [Google Scholar]
- 228.O'Connor, M. 2002. Insertions in the anticodon loop of tRNA1Gln(sufG) and tRNALys promote quadruplet decoding of CAAA. Nucleic Acids Res. 301985-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.O'Connor, M. 2002. Imbalance of tRNA(Pro) isoacceptors induces +1 frameshifting at near-cognate codons. Nucleic Acids Res. 30759-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.O'Connor, M. 2003. tRNA hopping: effects of mutant tRNAs. Biochim. Biophys. Acta 163041-46. [DOI] [PubMed] [Google Scholar]
- 231.O'Connor, M. 2007. Selection for intragenic suppressors of lethal 23S rRNA mutations in Escherichia coli identifies residues important for ribosome assembly and function. Mol. Genet. Genomics 278677-687. [DOI] [PubMed] [Google Scholar]
- 232.O'Connor, M., and A. E. Dahlberg. 1993. Mutations at U2555, a tRNA-protected base in 23S rRNA, affect translational fidelity. Proc. Natl. Acad. Sci. USA 909214-9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.O'Connor, M., and A. E. Dahlberg. 1995. The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J. Mol. Biol. 254838-847. [DOI] [PubMed] [Google Scholar]
- 234.O'Connor, M., and A. E. Dahlberg. 1996. The influence of base identity and base pairing on the function of the alpha-sarcin loop of 23S rRNA. Nucleic Acids Res. 242701-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.O'Connor, M., R. F. Gesteland, and J. F. Atkins. 1989. tRNA hopping: enhancement by an expanded anticodon. EMBO J. 84315-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.O'Connor, M., H. U. Göringer, and A. E. Dahlberg. 1992. A ribosomal ambiguity mutation in the 530 loop of E. coli 16S rRNA. Nucleic Acids Res. 204221-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.O'Connor, M., S. T. Gregory, and A. E. Dahlberg. 2004. Multiple defects in translation associated with altered ribosomal protein L4. Nucleic Acids Res. 325750-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.O'Connor, M., C. L. Thomas, R. A. Zimmermann, and A. E. Dahlberg. 1997. Decoding fidelity at the ribosomal A and P sites: influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res. 251185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.O'Connor, M., N. M. Willis, L. Bossi, R. F. Gesteland, and J. F. Atkins. 1993. Functional tRNAs with altered 3′ ends. EMBO J. 122559-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Öfverstedt, L. G., K. Zhang, S. Tapio, U. Skoglund, and L. A. Isaksson. 1994. Starvation in vivo for aminoacyl-tRNA increases the spatial separation between the two ribosomal subunits. Cell 79629-638. [DOI] [PubMed] [Google Scholar]
- 241.Ogle, J. M., D. E. Brodersen, W. M. Clemons, Jr., M. J. Tarry, A. P. Carter, and V. Ramakrishnan. 2001. Recognition of cognate transfer RNA by the 30s ribosomal subunit. Science 292897-902. [DOI] [PubMed] [Google Scholar]
- 242.Ohtsubo, Y., H. Genka, H. Komatsu, Y. Nagata, and M. Tsuda. 2005. High-temperature-induced transposition of insertion elements in Burkholderia multivorans ATCC 17616. Appl. Environ. Microbiol. 711822-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ohtsuki, T., T. Manabe, and M. Sisido. 2005. Multiple incorporation of non-natural amino acids into a single protein using tRNAs with non-standard structures. FEBS Lett. 5796769-6774. [DOI] [PubMed] [Google Scholar]
- 244.Olejniczak, M., and O. C. Uhlenbeck. 2006. tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie 88943-950. [DOI] [PubMed] [Google Scholar]
- 245.O'Mahony, D. J., D. Hughes, S. Thompson, and J. F. Atkins. 1989. Suppression of a −1 frameshift mutation by a recessive tRNA suppressor which causes doublet decoding. J. Bacteriol. 1713824-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.O'Mahony, D. J., B. H. Mims, S. Thompson, E. J. Murgola, and J. F. Atkins. 1989. Glycine tRNA mutants with normal anticodon loop size cause −1 frameshifting. Proc. Natl. Acad. Sci. USA 867979-7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Orgel, L. E. 1989. The origin of polynucleotide-directed protein synthesis. J. Mol. Evol. 29465-474. [DOI] [PubMed] [Google Scholar]
- 248.Orlova, M., A. Yueh, J. Leung, and S. P. Goff. 2003. Reverse transcriptase of Moloney murine leukemia virus binds to eukaryotic release factor 1 to modulate suppression of translational termination. Cell 115319-331. [DOI] [PubMed] [Google Scholar]
- 249.Pagel, F. T., T. M. F. Tuohy, J. F. Atkins, and E. J. Murgola. 1992. Doublet translocation at GGA is mediated directly by mutant transfer RNA2(Gly). J. Bacteriol. 1744179-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Pan, D., S. Kirillov, C. M. Zhang, Y. M. Hou, and B. S. Cooperman. 2006. Rapid ribosomal translocation depends on the conserved 18-55 base pair in P-site transfer RNA. Nat. Struct. Mol. Biol. 13354-359. [DOI] [PubMed] [Google Scholar]
- 251.Pedersen, S. 1984. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 32895-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Pestka, S. 1968. Studies on the formation of transfer ribonucleic acid-ribosome complexes. III. The formation of peptide bonds by ribosomes in the absence of supernatant enzymes. J. Biol. Chem. 2432810-2820. [PubMed] [Google Scholar]
- 253.Phelps, S. S., C. Gaudin, S. Yoshizawa, C. Benitez, D. Fourmy, and S. Joseph. 2006. Translocation of a tRNA with an extended anticodon through the ribosome. J. Mol. Biol. 369610-622. [DOI] [PubMed] [Google Scholar]
- 254.Pope, W. T., A. Brown, and R. H. Reeves. 1978. The identification of the tRNA substrates for the supK tRNA methylase. Nucleic Acids Res. 51041-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Pope, W. T., and R. H. Reeves. 1978. Purification and characterization of a tRNA methylase from Salmonella typhimurium. J. Bacteriol. 136191-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Prince, J. B., B. H. Taylor, D. L. Thurlow, J. Ofengand, and R. A. Zimmermann. 1982. Covalent crosslinking of tRNA1Val to 16S RNA at the ribosomal P site: identification of crosslinked residues. Proc. Natl. Acad. Sci. USA 795450-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Pulak, R., and P. Anderson. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 71885-1897. [DOI] [PubMed] [Google Scholar]
- 258.Qian, Q., and G. R. Björk. 1997. Structural alterations far from the anticodon of the tRNAGGGPro of Salmonella typhimurium induce +1 frameshifting at the peptidyl-site. J. Mol. Biol. 273978-992. [DOI] [PubMed] [Google Scholar]
- 259.Qian, Q., J. N. Li, H. Zhao, T. G. Hagervall, P. J. Farabaugh, and G. R. Björk. 1998. A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol. Cell 1471-482. [DOI] [PubMed] [Google Scholar]
- 260.Reeves, R. H., and J. R. Roth. 1971. A recessive UGA suppressor. J. Mol. Biol. 56523-533. [DOI] [PubMed] [Google Scholar]
- 261.Reeves, R. H., and J. R. Roth. 1975. Transfer ribonucleic acid methylase deficiency found in UGA supressor strains. J. Bacteriol. 124332-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Riddle, D. L., and J. Carbon. 1973. Frameshift suppression: a nucleotide addition in the anticodon of a glycine transfer RNA. Nature 242230-234. [DOI] [PubMed] [Google Scholar]
- 263.Riddle, D. L., and J. R. Roth. 1970. Suppressors of frameshift mutations in Salmonella typhimurium. J. Mol. Biol. 54131-144. [DOI] [PubMed] [Google Scholar]
- 264.Riddle, D. L., and J. R. Roth. 1972. Frameshift suppressors. II. Genetic mapping and dominance studies. J. Mol. Biol. 66483-493. [DOI] [PubMed] [Google Scholar]
- 265.Riddle, D. L., and J. R. Roth. 1972. Frameshift suppressors. III. Effects of suppressor mutations on transfer RNA. J. Mol. Biol. 66495-506. [DOI] [PubMed] [Google Scholar]
- 266.Riyasaty, S., and J. F. Atkins. 1968. External suppression of a frameshift mutant in Salmonella. J. Mol. Biol. 34541-557. [DOI] [PubMed] [Google Scholar]
- 267.Rodnina, M. V., A. Savelsbergh, V. I. Katunin, and W. Wintermeyer. 1997. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 38537-41. [DOI] [PubMed] [Google Scholar]
- 268.Rydén Aulin, M., and D. Hughes. 1990. Overproduction of release factor reduces spontaneous frameshifting and frameshift suppression by mutant elongation factor Tu. J. Bacteriol. 1726721-6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Sakai, H., R. Stiess, and B. Weiss-Brummer. 1991. Mitochondrial mutations restricting spontaneous translational frameshift suppression in the yeast Saccharomyces cerevisiae. Mol. Gen. Genetics 227306-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Samaha, R. R., R. Green, and H. F. Noller. 1995. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature 377309-314. [DOI] [PubMed] [Google Scholar]
- 271.Sambrook, J. F., D. P. Fan, and S. Brenner. 1967. A strong suppressor specific for UGA. Nature 214452-453. [DOI] [PubMed] [Google Scholar]
- 272.Sandbaken, M. G., and M. R. Culbertson. 1988. Mutations in elongation factor EF-1 alpha affect the frequency of frameshifting and amino acid misincorporation in Saccharomyces cerevisiae. Genetics 120923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Sanders, C. L., and J. F. Curran. 2007. Genetic analysis of the E site during RF2 programmed frameshifting. RNA 131483-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Sanders, C. L., K. J. Lohr, H. L. Gambill, R. B. Curran, and J. F. Curran. 2008. Anticodon loop mutations perturb reading frame maintenance by the E site tRNA. RNA 141874-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Sanderson, K. E., and J. A. Hurley. 1987. Linkage map of Salmonella typhimurium, p. 877-918. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. H. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, DC.
- 276.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 745463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Santer, U. V., J. Cekleniak, S. Kansil, M. Santer, M. O'Connor, and A. E. Dahlberg. 1995. A mutation at the universally conserved position 529 in Escherichia coli 16S rRNA creates a functional but highly error prone ribosome. RNA 189-94. [PMC free article] [PubMed] [Google Scholar]
- 278.Saulquin, X., E. Scotet, L. Trautmann, M. A. Peyrat, F. Halary, M. Bonneville, and E. Houssaint. 2002. +1 frameshifting as a novel mechanism to generate a cryptic cytotoxic T lymphocyte epitope derived from human interleukin 10. J. Exp. Med. 195353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Savelsbergh, A., V. I. Katunin, D. Mohr, F. Peske, M. V. Rodnina, and W. Wintermeyer. 2003. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol. Cell 111517-1523. [DOI] [PubMed] [Google Scholar]
- 280.Schwartz, R., and J. F. Curran. 1997. Analyses of frameshifting at UUU-pyrimidine sites. Nucleic Acids Res. 252005-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Seetharam, R., R. A. Heeren, E. Y. Wong, S. R. Braford, B. K. Klein, S. Aykent, C. E. Kotts, K. J. Mathis, B. F. Bishop, and M. J. Jennings. 1988. Mistranslation in IGF-1 during over-expression of the protein in Escherichia coli using a synthetic gene containing low frequency codons. Biochem. Biophys. Res. Commun. 155518-523. [DOI] [PubMed] [Google Scholar]
- 282.Selmer, M., C. M. Dunham, F. V. Murphy, A. Weixlbaumer, S. Petry, A. C. Kelley, J. R. Weir, and V. Ramakrishnan. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 3131935-1942. [DOI] [PubMed] [Google Scholar]
- 283.Sergiev, P. V., D. V. Lesnyak, S. V. Kiparisov, D. E. Burakovsky, A. A. Leonov, A. A. Bogdanov, R. Brimacombe, and O. A. Dontsova. 2005. Function of the ribosomal E-site: a mutagenesis study. Nucleic Acids Res. 336048-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Sharp, P. M., T. M. Tuohy, and K. R. Mosurski. 1986. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 145125-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Simonovic, M., and T. A. Steitz. 2008. Cross-crystal averaging reveals that the structure of the peptidyl-transferase center is the same in the 70S ribosome and the 50S subunit. Proc. Natl. Acad. Sci. USA 105500-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Simonson, A. B., and J. A. Lake. 2002. The transorientation hypothesis for codon recognition during protein synthesis. Nature 416281-285. [DOI] [PubMed] [Google Scholar]
- 287.Smith, W. S., H. Sierzputowska-Gracz, E. Sochacka, A. Malkiewicz, and P. F. Agris. 1992. Chemistry and structure of modified uridine dinucleosides are determined by thiolation. J. Am. Chem. Soc. 1147989-7997. [Google Scholar]
- 288.Sörensen, M. A., C. G. Kurland, and S. Pedersen. 1989. Codon usage determines translation rate in Escherichia coli. J. Mol. Biol. 207365-377. [DOI] [PubMed] [Google Scholar]
- 289.Sörensen, M. A., and S. Pedersen. 1991. Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J. Mol. Biol. 222265-280. [DOI] [PubMed] [Google Scholar]
- 290.Southworth, D. R., J. L. Brunelle, and R. Green. 2002. EFG-independent translocation of the mRNA: tRNA complex is promoted by modification of the ribosome with thiol-specific reagents. J. Mol. Biol. 324611-623. [DOI] [PubMed] [Google Scholar]
- 291.Spanjaard, R. A., and J. van Duin. 1988. Translation of the sequence AGG-AGG yields 50% ribosomal frameshift. Proc. Natl. Acad. Sci. USA 857967-7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Spirin, A. S. 1969. A model of the functioning ribosome: locking and unlocking of the ribosome subparticles. Cold Spring Harbor Symp. Quant. Biol. 39197-207. [DOI] [PubMed] [Google Scholar]
- 293.Spirin, A. S. 1985. Ribosomal translocation: facts and models. Prog. Nucleic Acid Res. Mol. Biol. 3275-114. [DOI] [PubMed] [Google Scholar]
- 294.Spirin, A. S. 1986. Ribosome structure and protein biosynthesis. The Benjamin/Cummings Publishing Company, Inc., Menlo Park, CA.
- 295.Sroga, G. E., F. Nemoto, Y. Kuchino, and G. R. Björk. 1992. Insertion (sufB) in the anticodon loop or base substitution (sufC) in the anticodon stem of tRNAPro2 from Salmonella typhimurium induces suppression of frameshift mutations. Nucleic Acids Res. 203463-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Stagg, S. M., M. Valle, R. K. Agrawal, J. Frank, and S. C. Harvey. 2002. Problems with the transorientation hypothesis. RNA 81093-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Stahl, G., S. Ben Salem, Z. Li, G. McCarty, A. Raman, M. Shah, and P. J. Farabaugh. 2001. Programmed +1 translational frameshifting in the yeast Saccharomyces cereviesiae results from disruption of translational error correction. Cold Spring Harbor Symp. Quant. Biol. 66249-258. [DOI] [PubMed] [Google Scholar]
- 298.Stahl, G., G. P. McCarty, and P. J. Farabaugh. 2002. Ribosome structure: revisiting the connection between translational accuracy and unconventional decoding. Trends Biochem. Sci. 27178-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Stahl, G., S. N. Salem, L. Chen, B. Zhao, and P. J. Farabaugh. 2004. Translational accuracy during exponential, postdiauxic, and stationary growth phases in Saccharomyces cerevisiae. Eukaryot. Cell 3331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Steitz, T. A. 2008. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 9242-253. [DOI] [PubMed] [Google Scholar]
- 301.Stent, G. S., and R. Calendar. 1978. Molecular genetics: an introductory narrative, 2nd ed. W. H. Freeman and Company, San Francisco, CA.
- 302.Sun, F. J., and G. Caetano-Anolles. 2008. Evolutionary patterns in the sequence and structure of transfer RNA: a window into early translation and the genetic code. PLoS One 3e2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Sundaram, M., P. C. Durant, and D. R. Davis. 2000. Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry 3912575-12584. [DOI] [PubMed] [Google Scholar]
- 304.Sundararajan, A., W. A. Michaud, Q. Qian, G. Stahl, and P. J. Farabaugh. 1999. Near-cognate peptidyl-tRNAs promote +1 programmed translational frameshifting in yeast. Mol. Cell 41005-1015. [DOI] [PubMed] [Google Scholar]
- 305.Taira, H., M. Fukushima, T. Hohsaka, and M. Sisido. 2005. Four-base codon-mediated incorporation of non-natural amino acids into proteins in a eukaryotic cell-free translation system. J. Biosci. Bioeng. 99473-476. [DOI] [PubMed] [Google Scholar]
- 306.Taira, H., T. Hohsaka, and M. Sisido. 2006. In vitro selection of tRNAs for efficient four-base decoding to incorporate non-natural amino acids into proteins in an Escherichia coli cell-free translation system. Nucleic Acids Res. 341653-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Taki, M., J. Matsushita, and M. Sisido. 2006. Expanding the genetic code in a mammalian cell line by the introduction of four-base codon/anticodon pairs. Chembiochem 7425-428. [DOI] [PubMed] [Google Scholar]
- 308.Taki, M., Y. Tokuda, T. Ohtsuki, and M. Sisido. 2006. Design of carrier tRNAs and selection of four-base codons for efficient incorporation of various nonnatural amino acids into proteins in Spodoptera frugiperda 21 (Sf21) insect cell-free translation system. J. Biosci. Bioeng. 102511-517. [DOI] [PubMed] [Google Scholar]
- 309.Tessier, P. M., and S. Lindquist. 2007. Prion recognition elements govern nucleation, strain specificity and species barriers. Nature 447556-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Thompson, J., D. F. Kim, M. O'Connor, K. R. Lieberman, M. A. Bayfield, S. T. Gregory, R. Green, H. F. Noller, and A. E. Dahlberg. 2001. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc. Natl. Acad. Sci. USA 989002-9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Topal, M. D., and J. R. Fresco. 1976. Complementary base pairing and the origin of substitution mutations. Nature 263285-289. [DOI] [PubMed] [Google Scholar]
- 312.Toulouse, A., F. Au-Yeung, C. Gaspar, J. Roussel, P. Dion, and G. A. Rouleau. 2005. Ribosomal frameshifting on MJD-1 transcripts with long CAG tracts. Hum. Mol. Genet. 142649-2660. [DOI] [PubMed] [Google Scholar]
- 313.Trifonov, E. N. 2004. The triplet code from first principles. J. Biomol. Struct. Dyn. 221-11. [DOI] [PubMed] [Google Scholar]
- 314.Tsuchihashi, Z., and P. O. Brown. 1992. Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNA(Lys) and an AAG lysine codon. Genes Dev. 6511-519. [DOI] [PubMed] [Google Scholar]
- 315.Tucker, S. D., E. J. Murgola, and F. T. Pagel. 1989. Missense and nonsense suppressors can correct frameshift mutations. Biochimie 71729-739. [DOI] [PubMed] [Google Scholar]
- 316.Tuohy, T. M., Z. Li, J. F. Atkins, and M. P. Deutscher. 1994. A functional mutant of tRNA(2Arg) with ten extra nucleotides in its TFC arm. J. Mol. Biol. 41369-1376. [DOI] [PubMed] [Google Scholar]
- 317.Tuohy, T. M., S. Thompson, R. F. Gesteland, and J. F. Atkins. 1992. Seven, eight and nine-membered anticodon loop mutants of tRNA2Arg which cause +1 frameshifting. Tolerance of DHU arm and other secondary mutations. J. Mol. Biol. 2281042-1054. [DOI] [PubMed] [Google Scholar]
- 318.Tuohy, T. M., S. Thompson, R. F. Gesteland, D. Hughes, and J. F. Atkins. 1990. The role of EF-Tu and other translation components in determining translocation step size. Biochim. Biophys. Acta 1050274-278. (Erratum, 1087:347.) [DOI] [PubMed] [Google Scholar]
- 319.Turner, D. H., and P. C. Bevilacqua. 1993. Thermodynamic considerations for evolution by RNA, p. 447-464. In R. F. Gesterland and J. F. Atkins (ed.), The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 320.Urbonavicius, J., Q. Qian, J. M. Durand, T. G. Hagervall, and G. R. Björk. 2001. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 204863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Urbonavicius, J., G. Stahl, J. M. Durand, S. N. Ben Salem, Q. Qian, P. J. Farabaugh, and G. R. Björk. 2003. Transfer RNA modifications that alter +1 frameshifting in general fail to affect −1 frameshifting. RNA 9760-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Varenne, S., J. Buc, R. Lloubes, and C. Lazdunski. 1984. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J. Mol. Biol. 180549-576. [DOI] [PubMed] [Google Scholar]
- 323.Vijgenboom, E., and L. Bosch. 1989. Translational frameshifts induced by mutant species of the polypeptide chain elongation factor Tu of Escherichia coli. J. Biol. Chem. 26413012-13017. [PubMed] [Google Scholar]
- 324.Waas, W. F., V. de Crécy-Lagard, and P. Schimmel. 2005. Discovery of a gene family critical to wyosine base formation in a subset of phenylalanine-specific transfer RNAs. J. Biol. Chem. 28037616-37622. [DOI] [PubMed] [Google Scholar]
- 325.Waas, W. F., Z. Druzina, M. Hanan, and P. Schimmel. 2007. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J. Biol. Chem. 28226026-26034. [DOI] [PubMed] [Google Scholar]
- 326.Walker, S. E., and K. Fredrick. 2006. Recognition and positioning of mRNA in the ribosome by tRNAs with expanded anticodons. J. Mol. Biol. 360599-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Wang, K., H. Neumann, S. Y. Peak-Chew, and J. W. Chin. 2007. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat. Biotechnol. 25770-777. [DOI] [PubMed] [Google Scholar]
- 328.Wang, L., J. Xie, and P. G. Schultz. 2006. Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 35225-249. [DOI] [PubMed] [Google Scholar]
- 329.Watson, J. D. 1964. The synthesis of proteins upon ribosomes. Bull. Soc. Chim. Biol. 461399-1425. [PubMed] [Google Scholar]
- 330.Watson, J. D. 1975. Molecular biology of the gene, 3rd ed. W. A. Benjamin, Inc., Menlo Park, CA.
- 331.Weiss, R., and J. Gallant. 1983. Mechanism of ribosome frameshifting during translation of the genetic code. Nature 302389-393. [DOI] [PubMed] [Google Scholar]
- 332.Weiss, R. B. 1984. Molecular model of ribosome frameshifting. Proc. Natl. Acad. Sci. USA 815797-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Weiss, R. B., D. M. Dunn, J. F. Atkins, and R. F. Gesteland. 1987. Slippery runs, shifty stops, backward steps, and forward hops: −2, −1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harbor Symp. Quant. Biol. 52687-693. [DOI] [PubMed] [Google Scholar]
- 334.Weiss, R. B., D. M. Dunn, J. F. Atkins, and R. F. Gesteland. 1990. Ribosomal frameshifting from −2 to +50 nucleotides. Prog. Nucleic Acid Res. Mol. Biol. 39159-183. [DOI] [PubMed] [Google Scholar]
- 335.Weiss, R. B., D. M. Dunn, M. Shuh, J. F. Atkins, and R. F. Gesteland. 1989. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1159-169. [PubMed] [Google Scholar]
- 336.Weiss, R. B., and J. A. Gallant. 1986. Frameshift suppression in aminoacyl-tRNA limited cells. Genetics 112727-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Weiss-Brummer, B., A. Zollner, A. Haid, and S. Thompson. 1995. Mutation of a highly conserved base in the yeast mitochondrial 21S rRNA restricts ribosomal frameshifting. Mol. Gen. Genetics 248207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Weixlbaumer, A., H. Jin, C. Neubauer, R. M. Voorhees, S. Petry, A. C. Kelley, and V. Ramakrishnan. 2008. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322953-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Wenthzel, A. M., M. Stancek, and L. A. Isaksson. 1998. Growth phase dependent stop codon readthrough and shift of translation reading frame in Escherichia coli. FEBS Lett. 421237-242. [DOI] [PubMed] [Google Scholar]
- 340.Whitfield, H. J., R. G. Martin, and B. N. Ames. 1966. Classification of aminotransferase (C gene) mutants in the histidine operon. J. Mol. Biol. 21335-355. [DOI] [PubMed] [Google Scholar]
- 341.Wickner, R. B. 1996. Prions and RNA viruses of Saccharomyces cerevisiae. Annu. Rev. Genet. 30109-139. [DOI] [PubMed] [Google Scholar]
- 342.Widerak, M., R. Kern, A. Malki, and G. Richarme. 2005. U2552 methylation at the ribosomal A-site is a negative modulator of translational accuracy. Gene 347109-114. [DOI] [PubMed] [Google Scholar]
- 343.Wills, N. M., and J. F. Atkins. 2006. The potential role of ribosomal frameshifting in generating aberrant proteins implicated in neurodegenerative diseases. RNA 121149-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Wills, N. M., M. O'Connor, C. C. Nelson, C. C. Rettberg, W. M. Huang, R. F. Gesteland, and J. F. Atkins. 2008. Translational bypassing without peptidyl-tRNA scanning of the coding gap mRNA. EMBO J. 272533-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Wilson, D. N., and K. H. Nierhaus. 2006. The E-site story: the importance of maintaining two tRNAs on the ribosome during protein synthesis. Cell. Mol. Life Sci. 632725-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Wilson, P. G., and M. R. Culbertson. 1988. SUF12 suppressor protein of yeast. A fusion protein related to the EF-1 family of elongation factors. J. Mol. Biol. 199559-573. [DOI] [PubMed] [Google Scholar]
- 347.Wimberly, B. T., D. E. Brodersen, W. M. Clemons, R. J. MorganWarren, A. P. Carter, C. Vonrhein, T. Hartsch, and V. Ramakrishnan. 2000. Structure of the 30S ribosomal subunit. Nature 407327-339. [DOI] [PubMed] [Google Scholar]
- 348.Woese, C. 1970. Molecular mechanics of translation: a reciprocating ratchet mechanism. Nature 226817-820. [DOI] [PubMed] [Google Scholar]
- 349.Woese, C. R. 2002. On the evolution of cells. Proc. Natl. Acad. Sci. USA 998742-8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Wolfe, M. D., F. Ahmed, G. M. Lacourciere, C. T. Lauhon, T. C. Stadtman, and T. J. Larson. 2004. Functional diversity of the rhodanese homology domain: the Escherichia coli ybbB gene encodes a selenophosphate-dependent tRNA 2-selenouridine synthase. J. Biol. Chem. 2791801-1809. [DOI] [PubMed] [Google Scholar]
- 351.Wong, J. T. 1991. Origin of genetically encoded protein synthesis: a model based on selection for RNA peptidation. Orig. Life Evol. Biosph. 21165-176. [DOI] [PubMed] [Google Scholar]
- 352.Xie, T., D. Ding, X. Tao, and D. Dafu. 1998. The relationship between synonymous codon usage and protein structure. FEBS Lett. 43493-96. [DOI] [PubMed] [Google Scholar]
- 353.Yamao, F., Y. Andachi, A. Muto, T. Ikemura, and S. Osawa. 1991. Levels of tRNAs in bacterial cells as affected by amino acid usage in proteins. Nucleic Acids Res. 196119-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Yourno, J. 1971. Similarity of cross-suppressible frameshifts in Salmonella typhimurium. J. Mol. Biol. 62223-231. [DOI] [PubMed] [Google Scholar]
- 355.Yourno, J. 1972. Externally suppressible +1 “glycine” frameshift: possible quadruplet isomers for glycine and proline. Nat. New Biol. 239219-221. [DOI] [PubMed] [Google Scholar]
- 356.Yourno, J., I. Ino, and T. Kono. 1971. A hotspot for spontaneous frameshift mutations in the histidinol dehydrogenase gene of Salmonella typhimurium. J. Mol. Biol. 62233-240. [DOI] [PubMed] [Google Scholar]
- 357.Yourno, J., and S. Tanemura. 1970. Restoration of in-phase translation by an unlinked suppressor of a frameshift mutation in Salmonella typhimurium. Nature 225422-426. [DOI] [PubMed] [Google Scholar]
- 358.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 A resolution. Science 292883-896. [DOI] [PubMed] [Google Scholar]
- 358a.Zaher, H. S., and R. Green. 2009. Quality control by the ribosome following peptide bond formation. Nature 457161-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Zavialov, A. V., V. V. Hauryliuk, and M. Ehrenberg. 2005. Guanine-nucleotide exchange on ribosome-bound elongation factor G initiates the translocation of tRNAs. J. Biol. 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Zhouravleva, G., L. Frolova, X. Legoff, R. Leguellec, S. Ingevechtomov, L. Kisselev, and M. Philippe. 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, erf1 and erf3. EMBO J. 144065-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Zinoni, F., A. Birkmann, T. C. Stadtman, and A. Böck. 1986. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc. Natl. Acad. Sci. USA 834650-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Zook, M. B., M. T. Howard, G. Sinnathamby, J. F. Atkins, and L. C. Eisenlohr. 2006. Epitopes derived by incidental translational frameshifting give rise to a protective CTL response. J. Immunol. 1766928-6934. [DOI] [PubMed] [Google Scholar]