Abstract
We developed a liquid bead microarray (LBMA) assay for genotyping genital human papillomaviruses (HPVs) based on the MY09-MY11-HMB01 PCR system and the reverse line blot (RLB) assay probe sequences. Using individual HPV plasmids, we were able to detect as few as 50 copies per reaction. In two separate retrospective studies, the LBMA assay was compared to the RLB assay and to the Hybrid Capture II (hc2) assay. Testing was performed without knowledge of other assay results. In the first study, 614 cervical swab samples (enriched for HPV infection) from 160 young women were tested for HPV DNA, and 360 (74.8%) type-specific HPV infections were detected by both assays, 71 (14.8%) by the LBMA assay only, and 50 (10.4%) by the RLB assay only. Type-specific agreement for the two assays was excellent (99.1%; kappa = 0.85; 95% confidence interval [95% CI], 0.82 to 0.88). Samples with discrepant LBMA and RLB test results tended to have low viral loads by a quantitative type-specific PCR assay. In the second study, cervical swab samples from 452 women (including 54 women with histologically confirmed cervical-intraepithelial neoplasia grade 2 or worse [≥CIN2]) were tested initially by the hc2 and subsequently by the LBMA assay. The estimated sensitivities for ≥CIN2 were similar for the LBMA and hc2 assays (98.4% [95% CI, 95.0 to 100%] and 95.6% [95% CI, 89.2 to 100%], respectively). The percentages of negative results among 398 women without ≥CIN2 were similar for the LBMA and hc2 assays (45% and 50%, respectively). The repeat test reproducibility for 100 samples was 99.1% (kappa = 0.92; 95% CI, 0.90 to 0.95). We conclude that the new LBMA assay will be useful for clinical and epidemiological research.
Human papillomavirus (HPV) is the central etiological agent for virtually all cervical cancers, for a substantial proportion of other anogenital tract cancers, and for a smaller proportion of head and neck cancers (3, 20). Currently, more than 100 different HPV types have been identified, and at least 40 types infect the anogenital epithelium. The risk of cancer is not the same for all HPV types. High-risk HPV types include HPV type 16 (HPV-16) and HPV-18, -31, -33, -35, -39, -45, -51, -52, -56, -58, -59, -68, -73, and -82. Low-risk HPV types include HPV-6, -11, -40, -42, -43, -44, -54, -61, -70, -72, -81, and -CP6108. Potentially high-risk types include HPV-26, -53, and -66 (20, 21). HPV DNA testing has been used (i) for triage of women with a Papanicolaou (Pap) test finding of atypical squamous cells of undetermined significance (ASC-US), (ii) for monitoring for recurrence of a precancerous cervical lesion or cancer after treatment, and (iii) as a primary screening method for cervical cancer in women 30 years old and older (1, 23, 31).
The only FDA-approved HPV assay for clinical testing, the Hybrid Capture II (hc2) assay, distinguishes high-risk HPVs from low-risk HPVs but does not provide individual HPV genotyping information. HPV type-specific assays are likely to have a role in the clinical management of the neoplastic diseases associated with HPV infection. Although 60 to 70% of U.S. women become infected with one or more high-risk genital types of HPV during their lifetime, most infections are quickly resolved and without consequence (2, 6, 19). Women who remain persistently positive for the same high-risk HPV type for extended periods are at increased risk for progression to cancer (11, 12). HPV genotyping is needed to differentiate women who are repeatedly positive for the same high-risk HPV type from those who are simply sequentially infected with different high-risk types of HPV. As the use of prophylactic HPV vaccines becomes more widespread, surveillance for population-level effectiveness will become an increasingly important activity that is likely to require the use of an HPV type-specific assay (10, 27). Estimates of the duration of vaccine-induced protection and the potential for herd immunity, for cross-protection, or for replacement (i.e., an increase in the prevalence of non-vaccine-type cervical lesions despite a decrease in the prevalence of vaccine-type lesions) will require HPV type-specific testing. Further, with increased coverage of HPV vaccines and the development of new vaccines that are likely to target more HPV types, the usefulness and methods of HPV testing and genotyping in cervical cancer prevention programs will likely need reevaluation and revision (14). Currently, HPV genotyping is indispensable for epidemiological and clinical studies of the transmission, natural history, and pathogenesis of HPV, and it is likely to have a role in the management of HPV-related precancerous lesions and cancers in the future.
DNA sequencing is the “gold standard” for HPV genotyping; however, it is costly, time-consuming, and difficult to apply to clinical samples, which frequently have multiple infections and produce nonspecific PCR products. Currently, the most widely used multiplex HPV genotyping assays are reverse line blot (RLB) assays (7, 15). These assays are based on solid-phase hybridization of amplified HPV sequences to a slot blot membrane. However, RLB assays are labor-intensive, are not easily automated, have limited reproducibility (because they rely on a subjective visual readout) (5), and are increasingly expensive.
Recent reports support the potential use of the liquid bead microarray (LBMA) assay based on Luminex technology for HPV genotyping, using either the existing GP5+-GP6+ system or the PGMY PCR system (13, 26, 28). This assay format is sensitive and amenable to high-throughput configuration and potentially can be automated. However, little is known about the analytical sensitivity and specificity of this new assay or about how it performs on clinical specimens in comparison to the other HPV genotyping assays.
In our present study, we developed an LBMA assay based on the MY09-MY11-HMB01 PCR system for genotyping clinically important HPV types (11, 18). We determined the analytical sensitivity and specificity of the LBMA assay using individual HPV plasmids, compared the genotyping results of the LBMA assay with those of the RLB assay, and estimated the clinical performance of the LBMA assay in comparison to that of the hc2 test using archived cervical swab samples.
MATERIALS AND METHODS
HPV LBMA assay development. (i) Probe selection.
The previously reported RLB probe sequences (Roche Molecular Diagnostics, Pleasanton, CA) were used in the development of our assay (7, 24). As in the RLB assay, two probes were used for HPV-16, -18, -31, -33, -35, -39, -42, -45, -51, -52, -53, -54, -55, -58, -59, -61, -62, -66, -67, -68, -69, -71, -72, -73, -81, -82, -83, -83, -IS39, and -CP6108, while a single probe was used for HPV-6, -11, -26, -40, -56, -64, and -70 (see the table in the supplemental material).
(ii) Probe conjugation to beads.
Thirty-seven Luminex bead sets (sets 17 to 21, 24, 25, 33 to 38, 42 to 47, 51 to 56, 61 to 66, and 72 to 77) were obtained (MiraiBio, Alameda, CA), and each bead set (approximately 5 × 106 beads/set) containing an intrinsic fluorescent signature was attached to specific HPV probes according to the manufacturer's oligonucleotide coupling protocol. Briefly, each bead set and its specific amine-substituted probes were first incubated with freshly prepared 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) at room temperature for 30 min and then washed and resuspended in Tris-EDTA at 50,000 beads/μl. All conjugated beads were pooled in suspension to form the 37-plex HPV-LBMA assay mixture (1,350 beads/set/μl).
HPV plasmids.
Plasmids of 12 HPV types (HPV-6, -11, -16, -18, -33, -35, -39, -45, -52, -59, -66, and -67) were available for the study. Nine of these (HPV-6, -11, -16, -33, -45, -52, -59, -66, and -67) contain full-length HPV genomes, while the remaining three (HPV-18, -35, and -39) contain a partial L1 open reading frame that encompasses the MY09-MY11-HMB01 amplicon (kindly donated by Denise Galloway). The concentrations of purified plasmids were determined with a UV spectrometer and were converted to copies per microliter. The plasmids were diluted, and 1 to 106 copies were mixed with 30 ng of purified K562 genomic DNA and amplified using MY09-MY11-HMB01 primers.
Amplification of the HPV L1 fragment.
MY09-MY11-HMB01 primers were used to amplify the HPV L1 fragment under the following conditions: 95°C for 9 min; 40 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 1 min; and a 5-min extension at 72°C. The ramp speed was set at 1°C/s (29). The presence of the correct PCR product was confirmed by gel electrophoresis; then the product was purified with a QIAquick PCR purification column (Qiagen, Valencia, CA) to remove the remaining excess primers.
Generation of a biotin-labeled single-stranded PCR product.
Biotin-labeled single-stranded HPV PCR products were generated from the PCR-amplified HPV L1 fragment by using biotin-labeled MY11 in place of MY09-MY11-HMB01 and the cycle conditions described above, except that only 20 cycles were run.
Hybridization assay.
One hundred nanograms of biotin-labeled PCR products or one-third of the second round of PCR products was mixed with conjugated beads, denatured at 95°C for 10 min, and hybridized at 55°C in 1.5× TMAC (tetramethyl ammonium chloride) solution for 30 min according to the Luminex oligonucleotide hybridization and DNA buffer protocols. After hybridization, the beads were washed twice with 1× TMAC using a 96-well filter plate (Qiagen, Valencia, CA) and then incubated with 4 μg/ml phycoerythrin-conjugated streptavidin (diluted in 1× TMAC; BD Pharmingen) at 55°C for 30 min. Finally, the beads were washed again with 1× TMAC and resuspended in 100 μl 1× TMAC for detection on the LiquiChip system (the Luminex 100 platform) according to the manufacturer's protocol (Qiagen, Valencia, CA). For each assay run, four HPV-negative controls (genomic DNA isolated from the K562 cell line) were included, and the background RLU was determined as the average for these four negative controls. A sample was considered to be weakly positive or positive for a specific HPV type if the RLU was greater than 7 or 10 times the background RLU, respectively, for that specific HPV.
HPV DNA load analysis by quantitative PCR.
Type-specific TaqMan assays were designed for HPV-6, -16, -39, -52, -53, -59, and -66 based on the HPV E7 gene. Primers and probe sequences are listed in Table 1. The quantitative PCR assays were performed on an ABI Prism 7900 sequence detection system (ABI, Foster City, CA). Relative quantification (RQ) was determined by the 2−ΔΔCT method. For each HPV type, the RQ of the sample with the highest viral load was set at 1.
TABLE 1.
Primers and probes for the quantification of HPV loads
| HPV type | Primer or probe (sequence)a |
|---|---|
| HPV-6 | F (GCAACGTTCGACTGGTTGTG) |
| Probe (6FAM-TGTACAGAAACAGACATCA-MGB) | |
| R (TCCCAACAGAAGCTGTTGCA) | |
| HPV-16 | F (CGGACAGAGCCCATTACAATATT) |
| Probe (6FAM-TAACCTTTTGTTGCAAGTGT-MGB) | |
| R (CGCACAACCGAAGCGTAGA) | |
| HPV-39 | F (ACCATGCAGTTAATCACCAACATC) |
| Probe (6FAM-ACTACTAGCCAGACGGG-MGB) | |
| R (TTGTGTGACGCTGTGGTTCA) | |
| HPV-52 | F (TGTGGACCGGCCAGATG) |
| Probe (6FAM-AGCAGAACAAGCCAC-MGB) | |
| R (CAACTGTGACAATATGTCACAATGTAGTAA) | |
| HPV-53 | F (GCAGTTGGCTGTTCAGAGTTCA) |
| Probe (6FAM-AAAGAGCTGCGTATTTT-MGB) | |
| R (TGTGCCCATAAGCATTTGTTG) | |
| HPV-59 | F (TGAAAAAGATGAACCAGATGGAGTT) |
| Probe (6FAM-ATCATCCTTTGCTACTAGCTA-MGB) | |
| R (TGTTGTGACGCTGTGGTTCAG) | |
| HPV-66 | F (GAGTTGGTGGTGCAGTTGGA) |
| Probe (6FAM-ATTCAGAGTACCAAAGAGG-MGB) | |
| R (AAGCAGCTGTTGTACCACACGTA) |
6FAM, 6-carboxyfluorescein; MGB, minor groove binder.
Clinical samples.
Archived cervical swab specimens collected in specimen transport medium (STM) from two different NIH-funded research projects were used in this study. Both sets of specimens were enriched for the detection of high-risk HPV infections. The first set of specimens consisted of 614 cervical swab samples from 160 female university students participating in a longitudinal study of the natural history of HPV infections (30). HPV genotyping by an RLB assay had been performed previously, and all RLB assay-positive cervical specimens, all RLB assay-negative cervical specimens for which a corresponding vulvar/vaginal or self-collected vaginal specimen was positive, and a random sample of the negative cervical specimens were included in this analysis. The second group of specimens consisted of cervical swab samples from 452 women enrolled in a cervical cancer screening study that included HPV DNA screening by the hc2 assay (16). The selected group of women was enriched for those with histologically confirmed diagnoses and included 54 women with cervical intraepithelial neoplasia grade 2 or worse (≥CIN2). The protease K-digested archived clinical samples, stored at −20°C, were used for reextraction via a QIAamp column according to the manufacturer's protocol (Qiagen, Valencia, CA) for the LBMA assay and the type-specific quantitative PCR assay. All LMBA assays were performed without knowledge of prior laboratory or clinical test results. The demographic characteristics of both study populations are listed in Table 2.
TABLE 2.
Characteristics of the women in the two study populations
| Population and characteristic | Value |
|---|---|
| Study population 1 | |
| No. of subjects | 160 |
| Age range (yr) (mean, SD) | 18-26 (21.9, 1.7) |
| No. (%) of the following race/ethnicity: | |
| White | 106 (66.3) |
| Asian | 30 (19.1) |
| Hispanic | 5 (2.9) |
| Other | 19 (11.7) |
| No. of cervical swab samples | 614 |
| No. (%) of samples with the following RLB assay result: | |
| Negative | 360 (58.6) |
| Positive | 254 (41.4) |
| No. (%) of positive samples with the following no. of HPV types detected: | |
| 1 types | 145 (57.1) |
| 2 types | 76 (29.9) |
| 3 types | 22 (8.7) |
| 4 types | 8 (3.2) |
| 5 types | 3 (1.2) |
| No. of persistent infections (no. of positive samples) | 107 (315) |
| No. of transient infections (no. of positive samples) | 95 (95) |
| Study population 2 | |
| No. of subjects | 452 |
| Age range (yr) (mean, SD) | 18-47 (24.1, 5.9) |
| No. (%) of the following race/ethnicity: | |
| White | 335 (74.1) |
| Black | 40 (8.8) |
| Hispanic | 20 (4.4) |
| Asian | 12 (2.7) |
| Other | 45 (10.0) |
| No. (%) with the following cervical hc2 assay result: | |
| Negative | 200 (44.2) |
| Positive | 252 (55.8) |
| No. (%) with the following histologic diagnosisa: | |
| Negative | 85 (39.5) |
| Atypia | 40 (18.6) |
| CIN1 | 30 (14.0) |
| CIN1 to CIN2 | 4 (1.9) |
| CIN2 | 17 (7.9) |
| CIN2 to CIN3 | 8 (3.7) |
| CIN3 or worse | 29 (13.5) |
| Insufficient | 2 (0.9) |
| No. (%) with the following liquid-based Pap test resultb: | |
| Negative | 295 (65.3) |
| ASC-US | 76 (16.8) |
| LSIL | 35 (7.7) |
| HSIL or greater | 37 (8.2) |
| Insufficient | 9 (2.0) |
A total of 215 subjects had histologically confirmed diagnoses.
LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion.
Statistical analysis.
The first group of archived cervical swab samples was used to assess the concordance of type-specific HPV DNA detection between the LBMA and RLB assays by using an unweighted kappa statistic to determine the percentage of agreement beyond that expected by chance. To account for correlation within subjects, 95% confidence intervals (95% CI) were computed using percentile bootstrap methods with 1,000 repetitions. By using data from the second group of cervical swab samples, different cervical cancer screening strategies were defined based on the use of LBMA, hc2, and/or cytology tests. Because only 13 HPV types (HPV-16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58, -59, and -68) are included in the high-risk hc2 assay, samples were considered positive for the LBMA assay only if 1 or more of those 13 HPV types were detected. Detection of other HPV types was considered a negative result for HPV for the purposes of this analysis. The strategies were evaluated for their sensitivities in detecting histologically confirmed ≥CIN2. Since this population was enriched for women with histologically confirmed disease, it was not possible to estimate meaningful measures of specificity for the four screening strategies based on the LBMA assay, the hc2 assay, and/or cytology. Instead, an indication of the relative specificities (and of the relative percentages of results that were “false positive” for ≥CIN2) of the LMBA and hc2 assays for a group of women without ≥CIN2 was provided by the calculated percentage of this group that is negative for high-risk HPV DNA by the LBMA or hc2 assay. Estimates of sensitivity were corrected for verification bias by using a previously described inverse probability weighting method (16). Ninety-five percent confidence intervals were computed using the 2.5th and 97.5th percentiles of the bootstrap distribution of weighted estimates of sensitivity. Weights were applied using the pweights command in Stata (StataCorp LP, TX). All statistical analyses were performed using STATA, version 10.0 (StataCorp LP, TX).
RESULTS
Analytical sensitivity and specificity of the LBMA assay.
We determined the detection limits of the assay using HPV plasmids. As shown in Table 3, the LBMA assay was able to detect as few as 50 copies of each HPV plasmid mixed with genomic DNA. The assay had high analytical specificity for each HPV type when tested with high copy numbers of plasmids (105 to 106 copies/reaction).
TABLE 3.
Detection of individual HPV plasmids by the LBMA assay
| Plasmid (no. of copies) | Result (RLU) with specific probes of the following type (cutoff)a:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPV-6 (358) | HPV-11 (293) | HPV-16 (263) | HPV-18 (393) | HPV-33 (367) | HPV-35 (322) | HPV-39 (473) | HPV-45 (418) | HPV-52 (317) | HPV-59 (405) | HPV-66 (312) | HPV-67 (331) | |
| Negative controls: | ||||||||||||
| dH2O | 17 | 18 | 23 | 26 | 24 | 15 | 17 | 24 | 20 | 28 | 27 | 28 |
| K562 | 27 | 18 | 17 | 35 | 27 | 29 | 44 | 34 | 25 | 36 | 24 | 37 |
| HPV-6 | ||||||||||||
| 95,722 | 1,560 | 23 | 24 | 31 | 25 | 22 | 28 | 35 | 25 | 29 | 28 | 28 |
| 100 | 763 | 26 | 26 | 41 | 28 | 29 | 35 | 33 | 19 | 32 | 22 | 37 |
| 50 | 361 | 32 | 29 | 38 | 29 | 26 | 32 | 32 | 21 | 31 | 27 | 38 |
| 10 | 735 | 23 | 24 | 31 | 25 | 27 | 29 | 34 | 23 | 34 | 26 | 34 |
| HPV-11 | ||||||||||||
| 348,786 | 17 | 1,515 | 10 | 18 | 19 | 17 | 17 | 18 | 14 | 23 | 12 | 28 |
| 100 | 14 | 580 | 10 | 21 | 13 | 14 | 22 | 21 | 11 | 26 | 18 | 28 |
| 50 | 14 | 532 | 13 | 20 | 20 | 14 | 23 | 25 | 13 | 27 | 17 | 30 |
| 10 | 19 | 145 | 16 | 27 | 21 | 15 | 21 | 27 | 16 | 27 | 23 | 29 |
| HPV-16 | ||||||||||||
| 106,476 | 28 | 31 | 1,145 | 38 | 41 | 32 | 32 | 54 | 25 | 40 | 30 | 39 |
| 100 | 45 | 41 | 1,991 | 68 | 45 | 43 | 43 | 56 | 45 | 50 | 44 | 51 |
| 50 | 39 | 33 | 2,004 | 36 | 41 | 38 | 35 | 46 | 35 | 48 | 36 | 47 |
| 10 | 32 | 27 | 1,733 | 32 | 41 | 40 | 41 | 41 | 25 | 44 | 33 | 43 |
| HPV-18 | ||||||||||||
| 1,187,373 | 24 | 23 | 27 | 2,098 | 25 | 32 | 30 | 28 | 27 | 33 | 26 | 33 |
| 100 | 27 | 26 | 18 | 2,359 | 21 | 24 | 34 | 32 | 16 | 38 | 26 | 41 |
| 50 | 32 | 30 | 21 | 1,740 | 24 | 26 | 30 | 35 | 25 | 29 | 25 | 37 |
| 10 | 30 | 25 | 26 | 41 | 29 | 20 | 31 | 40 | 26 | 34 | 24 | 38 |
| HPV-33 | ||||||||||||
| 25,395 | 29 | 35 | 19 | 35 | 2,499 | 28 | 29 | 38 | 24 | 39 | 33 | 43 |
| 100 | 26 | 24 | 28 | 35 | 2,608 | 31 | 28 | 33 | 21 | 38 | 30 | 34 |
| 50 | 26 | 21 | 18 | 26 | 2,660 | 25 | 18 | 32 | 18 | 32 | 26 | 32 |
| 10 | 22 | 17 | 21 | 27 | 2,658 | 18 | 27 | 26 | 20 | 34 | 28 | 29 |
| HPV-35 | ||||||||||||
| 568,041 | 33 | 19 | 36 | 30 | 27 | 2,310 | 27 | 33 | 23 | 39 | 30 | 42 |
| 100 | 42 | 33 | 31 | 43 | 37 | 2,566 | 41 | 52 | 31 | 50 | 34 | 45 |
| 50 | 35 | 37 | 29 | 47 | 34 | 2,158 | 43 | 50 | 32 | 46 | 37 | 55 |
| 10 | 35 | 36 | 35 | 38 | 35 | 122 | 37 | 46 | 36 | 43 | 34 | 45 |
| HPV-39 | ||||||||||||
| 20,766 | 33 | 36 | 32 | 44 | 17 | 28 | 1,678 | 36 | 30 | 34 | 31 | 43 |
| 100 | 15 | 14 | 14 | 22 | 15 | 16 | 449 | 20 | 12 | 22 | 15 | 21 |
| 50 | 17 | 13 | 11 | 23 | 15 | 16 | 490 | 22 | 13 | 24 | 17 | 21 |
| 10 | 13 | 12 | 12 | 21 | 12 | 14 | 50 | 21 | 11 | 24 | 17 | 22 |
| HPV-45 | ||||||||||||
| 311,229 | 38 | 29 | 30 | 35 | 20 | 24 | 30 | 782 | 32 | 39 | 31 | 38 |
| 100 | 28 | 26 | 22 | 31 | 29 | 30 | 27 | 2,044 | 21 | 32 | 30 | 31 |
| 50 | 26 | 16 | 19 | 37 | 26 | 25 | 26 | 1,991 | 22 | 34 | 28 | 32 |
| 10 | 20 | 23 | 22 | 29 | 28 | 23 | 29 | 40 | 29 | 33 | 27 | 43 |
| HPV-52 | ||||||||||||
| 476,139 | 31 | 26 | 28 | 29 | 35 | 34 | 31 | 43 | 1,616 | 38 | 32 | 36 |
| 100 | 39 | 39 | 34 | 54 | 33 | 28 | 36 | 46 | 1,108 | 41 | 36 | 53 |
| 50 | 45 | 41 | 43 | 55 | 50 | 41 | 46 | 51 | 722 | 51 | 40 | 50 |
| 10 | 29 | 30 | 26 | 39 | 27 | 22 | 24 | 38 | 27 | 31 | 26 | 36 |
| HPV-59 | ||||||||||||
| 102,762 | 44 | 29 | 39 | 45 | 43 | 38 | 36 | 39 | 44 | 1,534 | 45 | 46 |
| 100 | 29 | 25 | 27 | 43 | 32 | 19 | 30 | 40 | 23 | 1,099 | 21 | 34 |
| 50 | 27 | 31 | 24 | 30 | 30 | 18 | 30 | 35 | 28 | 1,065 | 21 | 34 |
| 10 | 15 | 21 | 12 | 23 | 22 | 13 | 26 | 24 | 20 | 210 | 24 | 27 |
| HPV-66 | ||||||||||||
| 107,706 | 19 | 23 | 22 | 38 | 28 | 15 | 23 | 33 | 18 | 24 | 2,121 | 37 |
| 100 | 27 | 23 | 16 | 29 | 29 | 29 | 22 | 28 | 23 | 36 | 1,655 | 36 |
| 50 | 24 | 24 | 20 | 25 | 24 | 25 | 32 | 29 | 16 | 34 | 1,266 | 32 |
| 10 | 23 | 21 | 17 | 30 | 25 | 22 | 26 | 25 | 20 | 30 | 1,585 | 37 |
| HPV-67 | ||||||||||||
| 248,838 | 25 | 15 | 12 | 27 | 134 | 18 | 28 | 26 | 19 | 30 | 21 | 1,703 |
| 100 | 36 | 20 | 11 | 22 | 20 | 18 | 46 | 34 | 16 | 33 | 20 | 1,082 |
| 50 | 22 | 15 | 14 | 23 | 26 | 17 | 37 | 27 | 15 | 31 | 16 | 1,259 |
| 10 | 21 | 14 | 12 | 16 | 22 | 12 | 28 | 26 | 9 | 24 | 14 | 517 |
Values for positive samples are in boldface. The cutoff for each HPV type is calculated as 10 times the mean RLU for 12 negative-control samples.
Agreement of the LBMA assay and the RLB assay.
The performance of the LBMA assay was evaluated using 614 archived cervical swab samples from 160 subjects. These specimens had previously been genotyped using the RLB assay for 27 HPV types. Because HPV-57 was not included in the LBMA assay, and there are four low-risk HPV types (HPV-40, -42, -54, and -55) that appear to be clinically insignificant, we restricted all comparisons to 22 genotypes (HPV-6, -11, -16, -18, -26, -31, -33, -35, -39, -45, -51, -52, -53, -56, -58, -59, -66, -68, -73, -82, -83, and -84). Of the 614 cervical swab samples, 254 (41.4%) were positive for one or more of these types of HPV by the RLB assay and 254 (41.4%) were positive by the LBMA assay. By the two assays together, a total of 481 type-specific HPV infections were detected. Overall, 74.8% of type-specific HPV infections were detected by both assays, 14.8% were detected by the LBMA assay only, and 10.4% were detected by the RLB assay only. By pooling across HPV types, the type-specific percentage of agreement for all HPV types was 99.1% (kappa = 0.85; 95% CI, 0.82 to 0.88) (Table 4).
TABLE 4.
HPV type-specific agreement on cervical swab samples
| HPV type | No. of results
|
Total | % Agreement | Kappa (95% CI) | |||
|---|---|---|---|---|---|---|---|
| RLB+ LBMA+ | RLB+ LBMA− | RLB− LBMA+ | RLB− LBMA− | ||||
| HPV-6 | 18 | 2 | 2 | 592 | 614 | 99.4 | 0.90 (0.78-0.98) |
| HPV-11 | 1 | 0 | 0 | 613 | 614 | 100 | 1 |
| HPV-16 | 55 | 2 | 5 | 552 | 614 | 98.9 | 0.93 (0.88, 0.97) |
| HPV-18 | 19 | 0 | 6 | 589 | 614 | 99.0 | 0.86 (0.74, 0.96) |
| HPV-26 | 0 | 0 | 9 | 605 | 614 | 98.5 | 0 |
| HPV-31 | 10 | 0 | 2 | 602 | 614 | 99.7 | 0.91 (0.75-1) |
| HPV-33 | 11 | 1 | 1 | 601 | 614 | 99.7 | 0.92 (0.76-1) |
| HPV-35 | 2 | 1 | 0 | 611 | 614 | 99.8 | 0.80 (0-1) |
| HPV-39 | 27 | 10 | 2 | 575 | 614 | 98.1 | 0.81 (0.67-0.90) |
| HPV-45 | 11 | 2 | 1 | 600 | 614 | 99.5 | 0.88 (0.71-1) |
| HPV-51 | 42 | 0 | 16 | 556 | 614 | 97.4 | 0.83 (0.74-0.90) |
| HPV-52 | 13 | 6 | 1 | 594 | 614 | 98.9 | 0.78 (0.60-0.93) |
| HPV-53 | 24 | 3 | 1 | 586 | 614 | 99.4 | 0.92 (0.82-0.98) |
| HPV-56 | 12 | 3 | 13 | 599 | 614 | 97.4 | 0.59 (0.39-0.75) |
| HPV-58 | 6 | 0 | 2 | 606 | 614 | 99.7 | 0.86 (0.56-1) |
| HPV-59 | 21 | 3 | 0 | 590 | 614 | 99.5 | 0.93 (0.84-1) |
| HPV-66 | 21 | 6 | 1 | 586 | 614 | 98.9 | 0.85 (0.72-0.94) |
| HPV-68 | 2 | 0 | 0 | 612 | 614 | 100 | 1 |
| HPV-73 | 7 | 1 | 0 | 606 | 614 | 99.8 | 0.93 (0.75-1) |
| HPV-82 | 9 | 0 | 2 | 603 | 614 | 99.7 | 0.90 (.71-1) |
| HPV-83 | 6 | 0 | 0 | 608 | 614 | 100 | 1 |
| HPV-84 | 43 | 10 | 7 | 554 | 614 | 97.2 | 0.82 (0.73-0.90) |
| All types | 360 | 50 | 71 | 13,027 | 13,508 | 99.1 | 0.85 (0.82-0.88) |
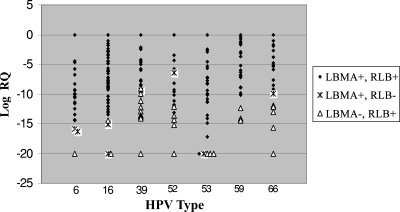
In order to further understand the cause of the discrepancy between the two assays, type-specific quantitative PCR assays targeting the HPV E7 region were designed. Specifically, we focused on high-risk or potentially high risk HPV types (HPV-16, -39, -52, -53, -59, and -66) for which the RLB assay gave positive results but that were more likely missed by the LBMA assay. In addition, an HPV-6 type-specific assay was designed. We analyzed samples that were positive by either the RLB or the LBMA assay for one or more of these seven HPV types. Samples showing discordant results between the RLB and the LBMA assays tended to have lower viral loads than samples that were positive by both assays (Fig. 1).
FIG. 1.
HPV loads of samples positive by either the LBMA or the RLB assay. DNA viral loads of samples positive for HPV-6, -16, -39, -52, -53, -59, or -66 by either the LBMA or the RLB assay were determined by a quantitative PCR assay based on the HPV E7 sequence. RQ was determined using the 2−ΔΔCT method. For each HPV type, the RQ of the sample with the highest viral load was set at 1.
Relative performances of the LBMA and hc2 assays for detection of histologically confirmed ≥CIN2.
The performance of screening strategies defined by results from the LBMA assay, the hc2 assay, and/or cytology was evaluated for 452 women (Table 5). Among 54 women with histologically confirmed ≥CIN2, the estimated weighted sensitivities for the LBMA and hc2 assays were similar (98.4% [95% CI, 95.0 to 100] and 95.6% [95% CI, 89.2 to 100], respectively). The percentages of negative results for 398 women without histologically confirmed ≥CIN2 were also similar for the LBMA and RLB assays (45% and 50%, respectively) (Table 6).
TABLE 5.
Sensitivities of four different strategies for detecting histologically confirmed ≥CIN2a
| Strategy | Criterion for referral to colposcopy | Weighted sensitivity (%) (95% CI) |
|---|---|---|
| LBMA assay for high-risk HPV DNA | Positive LBMA result for at least 1 of 13 high-risk HPV types targeted by the hc2 assayb | 98.4 (95.0-100) |
| hc2 assay for high-risk HPV DNA | Positive hc2 assay result for high-risk HPV DNA | 95.6 (89.2-100) |
| Pap testing with reflex hc2 testing for ASC-US | Liquid-based cytology result of ≥LSIL,c or liquid-based cytology result of ASC-US and positive hc2 assay result for high-risk HPV DNA | 72.7 (60.2-85.5) |
| Pap testing with reflex LBMA testing for ASC-US | Liquid-based cytology result of ≥LSIL, or liquid-based cytology result of ASC-US and positive LBMA result for at least 1 of 13 high-risk HPV types targeted by the hc2 assay | 74.8 (62.4-86.2) |
Insufficient and inadequate results were included in the “positive screening result” category, and for the strategy involving Pap testing with reflex LBMA testing for ASC-US, missing follow-up hc2 assay results were considered to be negative. Changing these assumptions did not markedly impact the findings.
High-risk HPV types not targeted by the hc2 assay were included in the “negative for HPV DNA” category.
≥LSIL, low-grade squamous intraepithelial lesion or worse. Inadequate cytology results were included in the ≥LSIL category, since women with inadequate cytology results are generally asked to return for a follow-up visit.
TABLE 6.
LBMA and hc2 test results for women with and without histologically confirmed ≥CIN2
| Histologic categorya | No. (%) positive by the following assay:
|
|
|---|---|---|
| LBMAb | hc2 | |
| Histologically confirmed (n = 213) | ||
| ≥CIN2 (n = 54) | 53 (98.2) | 52 (96.2) |
| ≥CIN3 (n = 37) | 36 (97.3) | 36 (97.3) |
| CIN2 (n = 17) | 17 (100.0) | 16 (94.1) |
| CIN1 or less (n = 159) | 124 (78.0) | 121 (76.1) |
| No histological confirmation (n = 239) | ||
| Cytologic findings of SIL (n = 13) | 12 (92.3) | 13 (100.0) |
| Cytologic findings of ASC-US (n = 26) | 14 (53.9) | 14 (53.9) |
| Normal cytologic findings (n = 197) | 67 (34.0) | 52 (26.4) |
| Inadequate cytologic findings (n = 3) | 1 (33.3) | 0 (0.0) |
SIL, squamous intraepithelial lesion.
Number (percent) positive for at least 1 of 13 high-risk HPV types targeted by the hc2 assay. High-risk types not targeted by the hc2 assay were included in the “negative for HPV DNA” category.
Reproducibility of the LBMA assay.
We repeated the LBMA assay on 100 randomly selected cervical swab samples. When each of 37 HPV types was considered separately for the reproducibility analysis (100 × 37 = 3,700 comparisons in total), 207 (5.6%) samples were concordantly positive, 3,460 (93.5%) were concordantly negative, and 33 (0.9%) were discordant. Therefore the agreement between the two repeated tests was 99.1% (kappa = 0.92; 95% CI, 0.90 to 0.95).
DISCUSSION
We developed an LBMA assay for genotyping HPVs. The LBMA assay was able to detect as few as 50 copies of the HPV genome and displayed high analytical specificity. When tested on 614 archived cervical samples, the LBMA assay showed excellent reproducibility and excellent agreement with the RLB assay for HPV genotyping. Using cervical swab samples from 452 subjects, we observed that the LBMA assay had an estimated clinical sensitivity for ≥CIN2 that was comparable to that of the hc2 assay. These results indicate that this newly developed LBMA assay is likely to be a valid and reliable alternative method for HPV genotyping and a sensitive assay for identifying high-grade cervical lesions.
Several previous studies reported the feasibility of establishing a Luminex-based HPV genotyping assay targeting 15 to 45 different HPVs (9, 13, 17, 22, 26, 28). Various primer systems were used for the development of these Luminex assays, including PGMY09-PGMY11 (28), GP5+-GP6+ (26), MY09-MY11 (13), type-specific primers (9), and YBT L1-GP6-1 (22). Using archived clinical samples, these assays reported 74 to 99% agreement with other HPV detection (hc2 assay) or genotyping (RLB assay, type-specific PCR, or HPV microarray assay) assays (Table 7).
TABLE 7.
Comparison of various reported bead-based HPV genotyping assays
| Primer | No. of HPV types detected | No. of clinical samples | Assay compared | Reported agreement (%) | Reported kappa (95% CI) | Source or reference |
|---|---|---|---|---|---|---|
| PGMY09-PGMY11 | 45 | 429 | hc2 | 82 | 0.68 (0.65, 0.72) | 28 |
| GP5+-GP6+ | 22 | 94 | RLB | 85 | 0.92 | 26 |
| MY09-MY11 | 26 | 133 | Type-specific PCR sequencing | 90 | 13 | |
| Type specific | 25 | 109 | hc2 | 95 | 9 | |
| YBT L1-GP6-1 | 15 | 53 | HPV microarray | 74 | 22 | |
| MY09-MY11-HMB01 | 22 | 614 | RLB | 99 | 0.85 (0.82, 0.88) | Present study |
While these previous reports support the LBMA technology for HPV DNA genotyping, to our knowledge, the present report is the first to provide results of a large-scale investigation that compared HPV genotyping results for the LBMA assay to those of widely used HPV genotyping assays based on RLB technology. In addition, the results for the LBMA assay were compared to those of the FDA-approved hc2 test in order to assess the potential clinical performance of the LBMA assay for detection of ≥CIN2. Although many of the samples included in the genotyping comparison were positive for more than 1 of the 22 targeted HPV types (42.9% of 254 RLB assay-positive samples), type-specific agreement was high, indicating excellent sensitivity and specificity for type-specific HPV DNA detection, even in the setting of mixed infections. In the current study, we did not compare the LBMA assay to the commercially available Roche Linear Array (LA) assay, because these archived samples had already been genotyped by the RLB assay. Recently, Castle et al. compared the performances of the RLB and LA assays using archived clinical samples (3). The percentage of agreement for carcinogenic HPV DNA detection was 88% (kappa = 0.76). Although the LA assay appeared to be more sensitive than the RLB assay in detecting HPV DNA, the authors suggested that this was due largely to the different DNA isolation methods used and the different amounts of DNA input for PCR amplification. When equal amounts of DNA extracted by the same method were used, no difference in overall or type-specific HPV detection was observed (3). The LBMA assay, like any other bead-based assay or solid-phase assay with an automated reading, is possibly more objective than the RLB assay, because unlike the RLB assay, it does not rely on a subjective visual readout. Moreover, the LMBA assay is amenable to a high-throughput configuration, can potentially be automated, can easily be scaled up to 100 HPV types, and can be combined with Luminex-based assays for the detection of HPV type-specific antibodies.
Several aspects of the LBMA assay might be improved in the future. First, a β-globin or other housekeeping gene probe could be incorporated to monitor the sample input. Second, the assay could be streamlined by using fluorescently labeled primers in the PCR, to avoid the need for a second hybridization with phycoerythrin-conjugated streptavidin. Third, the assay sensitivity could be improved by using biotin- or fluorescent dye-labeled nucleotides in the PCR instead of labeled primers. Finally, in order to expand the LBMA assay to allow it to genotype cutaneous HPV types, which are more heterogeneous and for which it is difficult to design common degenerate primers for amplification, signal amplification should be considered as an alternative to PCR amplification.
In conclusion, our data showed excellent correlation between the RLB and the LBMA assays when they were used for genotyping clinical samples, and it also showed comparable sensitivities for the hc2 and LMBA assays for the detection of biopsy-confirmed CIN2 or worse. Several recent studies underscore the importance of standardization of HPV genotyping assay protocols (3-5, 8, 25). The LBMA assay described here is amenable to standardization and thus shows promise for use in large-scale epidemiological studies of HPV pathogenesis, in surveillance of HPV immunization programs for population-level effectiveness, and in clinical investigations of new approaches to the prevention, diagnosis, and management of HPV-related cancers and precancerous lesions.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Health/National Cancer Institute (CA34493 and CA105181).
Informed consent was obtained according to procedures approved by the Human Subjects Committee of the University of Washington.
Footnotes
Published ahead of print on 14 January 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.ASCUS-LSIL Triage Study (ALTS) Group. 2003. Results of a randomized trial on the management of cytology interpretations of atypical squamous cells of undetermined significance. Am. J. Obstet. Gynecol. 1881383-1392. [DOI] [PubMed] [Google Scholar]
- 2.Baseman, J. G., and L. A. Koutsky. 2005. The epidemiology of human papillomavirus infections. J. Clin. Virol. 32(Suppl. 1)S16-S24. [DOI] [PubMed] [Google Scholar]
- 3.Castle, P. E., P. E. Gravitt, D. Solomon, C. M. Wheeler, and M. Schiffman. 2008. Comparison of linear array and line blot assay for detection of human papillomavirus and diagnosis of cervical precancer and cancer in the Atypical Squamous Cell of Undetermined Significance and Low-Grade Squamous Intraepithelial Lesion Triage Study. J. Clin. Microbiol. 46109-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castle, P. E., C. Porras, W. G. Quint, A. C. Rodriguez, M. Schiffman, P. E. Gravitt, P. Gonzalez, H. A. Katki, S. Silva, E. Freer, L.-J. Van Doorn, S. Jiménez, R. Herrero, and A. Hildesheim for the CVT Group. 2008. Comparison of two PCR-based human papillomavirus genotyping methods. J. Clin. Microbiol. 463437-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn, S. T., R. A. Allen, S. Wang, J. Walker, and M. Schiffman. 2007. DNA extraction: an understudied and important aspect of HPV genotyping using PCR-based methods. J. Virol. Methods 14345-54. [DOI] [PubMed] [Google Scholar]
- 6.Gravitt, P. E., and R. Jamshidi. 2005. Diagnosis and management of oncogenic cervical human papillomavirus infection. Infect. Dis. Clin. N. Am. 19439-458. [DOI] [PubMed] [Google Scholar]
- 7.Gravitt, P. E., C. L. Peyton, R. J. Apple, and C. M. Wheeler. 1998. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J. Clin. Microbiol. 363020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gravitt, P. E., M. Schiffman, D. Solomon, C. M. Wheeler, and P. E. Castle. 2008. A comparison of linear array and hybrid capture 2 for detection of carcinogenic human papillomavirus and cervical precancer in ASCUS-LSIL triage study. Cancer Epidemiol. Biomarkers Prev. 171248-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han, J., D. C. Swan, S. J. Smith, S. H. Lum, S. E. Sefers, E. R. Unger, and Y. W. Tang. 2006. Simultaneous amplification and identification of 25 human papillomavirus types with Templex technology. J. Clin. Microbiol. 444157-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harper, D. M., E. L. Franco, C. Wheeler, D. G. Ferris, D. Jenkins, A. Schuind, T. Zahaf, B. Innis, P. Naud, N. S. De Carvalho, C. M. Roteli-Martins, J. Teixeira, M. M. Blatter, A. P. Korn, W. Quint, and G. Dubin. 2004. Efficacy of a bivalent L1 virus-like particle vaccine in prevention of infection with human papillomavirus types 16 and 18 in young women: a randomised controlled trial. Lancet 3641757-1765. [DOI] [PubMed] [Google Scholar]
- 11.Hildesheim, A., M. H. Schiffman, P. E. Gravitt, A. G. Glass, C. E. Greer, T. Zhang, D. R. Scott, B. B. Rush, P. Lawler, M. E. Sherman, et al. 1994. Persistence of type-specific human papillomavirus infection among cytologically normal women. J. Infect. Dis. 169235-240. [DOI] [PubMed] [Google Scholar]
- 12.Ho, G. Y., R. Bierman, L. Beardsley, C. J. Chang, and R. D. Burk. 1998. Natural history of cervicovaginal papillomavirus infection in young women. N. Engl. J. Med. 338423-428. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, H. L., H. H. Zhu, L. F. Zhou, F. Chen, and Z. Chen. 2006. Genotyping of human papillomavirus in cervical lesions by L1 consensus PCR and the Luminex xMAP system. J. Med. Microbiol. 55715-720. [DOI] [PubMed] [Google Scholar]
- 14.Kiviat, N. B., S. E. Hawes, and Q. Feng. 2008. Screening for cervical cancer in the era of the HPV vaccine—the urgent need for both new screening guidelines and new biomarkers. J. Natl. Cancer Inst. 100290-291. [DOI] [PubMed] [Google Scholar]
- 15.Kleter, B., L. J. van Doorn, L. Schrauwen, A. Molijn, S. Sastrowijoto, J. ter Schegget, J. Lindeman, B. ter Harmsel, M. Burger, and W. Quint. 1999. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J. Clin. Microbiol. 372508-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulasingam, S. L., J. P. Hughes, N. B. Kiviat, C. Mao, N. S. Weiss, J. M. Kuypers, and L. A. Koutsky. 2002. Evaluation of human papillomavirus testing in primary screening for cervical abnormalities: comparison of sensitivity, specificity, and frequency of referral. JAMA 2881749-1757. [DOI] [PubMed] [Google Scholar]
- 17.Liu, M., C. X. Wang, X. M. Deng, L. S. Wang, J. Zhang, W. Li, G. X. Zheng, and J. F. Wang. 2007. Study on the genotyping of human papillomavirus using a new DNA liquid chip in women of high-risk group of Shandong province. Zhonghua Liu Xing Bing Xue Za Zhi. 28487-490. (In Chinese.) [PubMed] [Google Scholar]
- 18.Manos, M. M., Y. Ting, D. K. Wright, A. J. Lewis, T. R. Broker, and S. M. Wolinsky. 1989. The use of polymerase chain reaction amplification for the detection of genital human papillomavirus, p. 209-214. In M. Furth and M. Greaves (ed.), Molecular diagnostics of human cancer, vol. 7. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 19.Monsonego, J., F. X. Bosch, P. Coursaget, J. T. Cox, E. Franco, I. Frazer, R. Sankaranarayanan, J. Schiller, A. Singer, T. C. Wright, Jr., W. Kinney, C. J. Meijer, J. Linder, E. McGoogan, and C. Meijer. 2004. Cervical cancer control, priorities and new directions. Int. J. Cancer 108329-333. [DOI] [PubMed] [Google Scholar]
- 20.Muñoz, N., F. X. Bosch, S. de Sanjose, R. Herrero, X. Castellsagué, K. V. Shah, P. J. Snijders, and C. J. Meijer. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348518-527. [DOI] [PubMed] [Google Scholar]
- 21.Muñoz, N., X. Castellsagué, A. B. de González, and L. Gissmann. 2006. Chapter 1: HPV in the etiology of human cancer. Vaccine 24(Suppl. 3)S1-S10. [DOI] [PubMed] [Google Scholar]
- 22.Oh, Y., S. M. Bae, Y. W. Kim, H. S. Choi, G. H. Nam, S. J. Han, C. H. Park, Y. Cho, B. D. Han, and W. S. Ahn. 2007. Polymerase chain reaction-based fluorescent Luminex assay to detect the presence of human papillomavirus types. Cancer Sci. 98549-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paraskevaidis, E., M. Arbyn, A. Sotiriadis, E. Diakomanolis, P. Martin-Hirsch, G. Koliopoulos, G. Makrydimas, J. Tofoski, and D. H. Roukos. 2004. The role of HPV DNA testing in the follow-up period after treatment for CIN: a systematic review of the literature. Cancer Treat. Rev. 30205-211. [DOI] [PubMed] [Google Scholar]
- 24.Peyton, C. L., P. E. Gravitt, W. C. Hunt, R. S. Hundley, M. Zhao, R. J. Apple, and C. M. Wheeler. 2001. Determinants of genital human papillomavirus detection in a US population. J. Infect. Dis. 1831554-1564. [DOI] [PubMed] [Google Scholar]
- 25.Sabol, I., M. Salakova, J. Smahelova, M. Pawlita, M. Schmitt, N. M. Gasperov, M. Grce, and R. Tachezy. 2008. Evaluation of different techniques for identification of human papillomavirus types of low prevalence. J. Clin. Microbiol. 461606-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitt, M., I. G. Bravo, P. J. Snijders, L. Gissmann, M. Pawlita, and T. Waterboer. 2006. Bead-based multiplex genotyping of human papillomaviruses. J. Clin. Microbiol. 44504-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villa, L. L., R. L. Costa, C. A. Petta, R. P. Andrade, K. A. Ault, A. R. Giuliano, C. M. Wheeler, L. A. Koutsky, C. Malm, M. Lehtinen, F. E. Skjeldestad, S. E. Olsson, M. Steinwall, D. R. Brown, R. J. Kurman, B. M. Ronnett, M. H. Stoler, A. Ferenczy, D. M. Harper, G. M. Tamms, J. Yu, L. Lupinacci, R. Railkar, F. J. Taddeo, K. U. Jansen, M. T. Esser, H. L. Sings, A. J. Saah, and E. Barr. 2005. Prophylactic quadrivalent human papillomavirus (types 6, 11, 16, and 18) L1 virus-like particle vaccine in young women: a randomised double-blind placebo-controlled multicentre phase II efficacy trial. Lancet Oncol. 6271-278. [DOI] [PubMed] [Google Scholar]
- 28.Wallace, J., B. A. Woda, and G. Pihan. 2005. Facile, comprehensive, high-throughput genotyping of human genital papillomaviruses using spectrally addressable liquid bead microarrays. J. Mol. Diagn. 772-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weaver, B. A., Q. Feng, K. K. Holmes, N. Kiviat, S. K. Lee, C. Meyer, M. Stern, and L. A. Koutsky. 2004. Evaluation of genital sites and sampling techniques for detection of human papillomavirus DNA in men. J. Infect. Dis. 189677-685. [DOI] [PubMed] [Google Scholar]
- 30.Winer, R. L., J. P. Hughes, Q. Feng, S. O'Reilly, N. B. Kiviat, K. K. Holmes, and L. A. Koutsky. 2006. Condom use and the risk of genital human papillomavirus infection in young women. N. Engl. J. Med. 3542645-2654. [DOI] [PubMed] [Google Scholar]
- 31.Wright, T. C., Jr., M. Schiffman, D. Solomon, J. T. Cox, F. Garcia, S. Goldie, K. Hatch, K. L. Noller, N. Roach, C. Runowicz, and D. Saslow. 2004. Interim guidance for the use of human papillomavirus DNA testing as an adjunct to cervical cytology for screening. Obstet. Gynecol. 103304-309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.