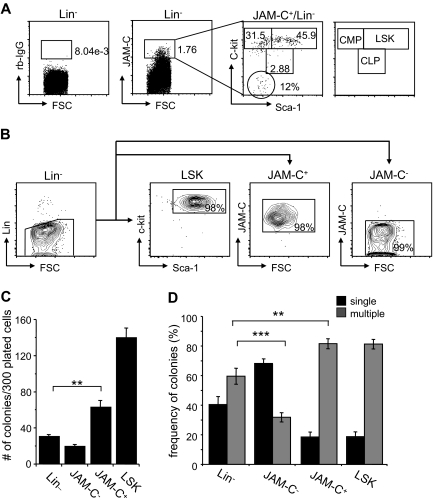
Figure 2.
Colony formation potential of JAM-C–expressing BM cells. (A) BM cells were stained with anti-lineage cocktail (anti-CD3ϵ, anti-B220, anti-CD11b, anti–Gr-1, and anti-Ter119), anti–Sca-1, anti–c-Kit, and anti–JAM-C. Subsequently, the Lin− cells were sorted into populations lacking JAM-C expression (JAM-C−), expressing JAM-C (JAM-C+), or expressing Sca-1 and c-Kit (LSK). Numbers indicate the percentage of cells in each gate after the sort. Shown is a representative FACS sort profile. (B) JAM-C+Lin− cells were gated and analyzed for expression of c-Kit and Sca-1, which are differentially expressed on hematopoietic progenitors (shown in the far right panel). Staining with control rabbit serum IgG is shown on the left of the dot plot. Numbers indicate the percentage of cells in each gate. (C) Lin−, JAM-C−, JAM-C+, and LSK cells were sorted from BM at a density of one cell per well into 96-well plates containing IMDM with mSCF, mIL-3, mIL-6, and hEPO. After 10 days of culture, the number of colonies was determined. Data represents mean number of colonies plus or minus SEM that grew in a total of 300 wells in 4 independent experiments. (D) Lin−, JAM-C−, JAM-C+, and LSK cells were sorted from BM for analysis of in vitro colony-forming ability. A total of 1000 Lin− and JAM-C− cells or 200 JAM-C+ and LSK cells were plated in methylcellulose containing mSCF, mIL-3, mIL-6, and hEPO. After 10 days, colonies were assigned scores for the presence of colonies containing single lineages (■) or multiple lineages (■). Data represent mean plus or minus SEM on duplicate plates of 4 independent experiments. Significances are shown on the graph. **P ≤ .01; ***P ≤ .001.