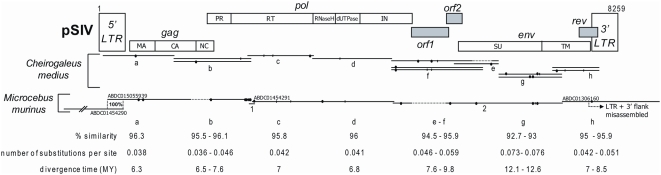
Figure 1. Map of the consensus pSIV reconstructed in this study based on one full-length copy of pSIVgml (Microcebus murinus) and between one and three copies of pSIVfdl (Cheirogaleus medius) (the alignment is provided in Dataset S1).
The LTR fragments contained in the ABDC01505939 and ABDC01454290 contigs correspond to the same full length pSIV copy (see Figure S1). The contig ABDC01306160 results from a misassembly between a trace read containing a solo-LTR and a trace read containing the 3′ terminus of the env of the full-length pSIVgml and a fragment of the 3′ LTR (see Figure S1). The different domains of pSIV were identified by comparison with the HIV1-HXB2 reference sequence [40] (see Figure S4 for a more precise map). Closed circles: non-sense frameshifts. Vertical bars: in frame stop codons. Dash lines: missing fragments. The range of pairwise similarity, number of substitutions per site and inferred divergence times between pSIVgml and pSIVfdl sequences are indicated.