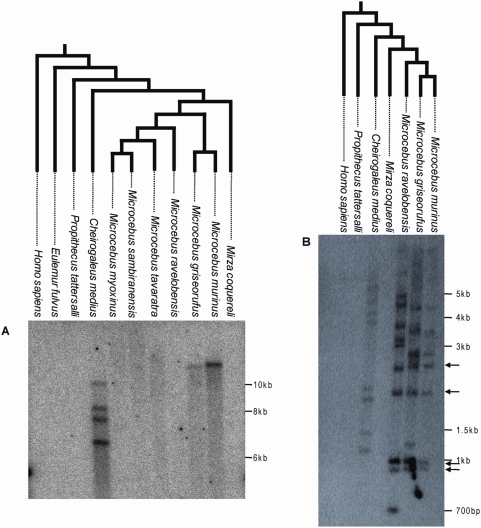
Figure 2. Southern blot of digested genomic DNA of various Malagasy lemurs and human using a ∼1 kb probe corresponding to a fragment of pSIVgml env (A) or a ∼300 bp probe corresponding to a fragment of the pSIV LTR (B).
Arrows highlight bands of the same size shared by Microcebus murinus, M. griseorufus and M. ravelobensis but not Cheirogaleus medius. These bands likely correspond to solo-LTRs located at orthologous position in the three Microcebus species. The trees on top of each blot depict the phylogenetic relationships of the species according to [23],[41]. See Table S4 for the voucher specimen numbers of the lemur tissue samples used in this study. A picture of the ethidium bromide stained gels used to prepare the blots is shown in Figure S2. The absence of pSIV in Mirza was confirmed by PCR using different sets of primers (Figure S3).