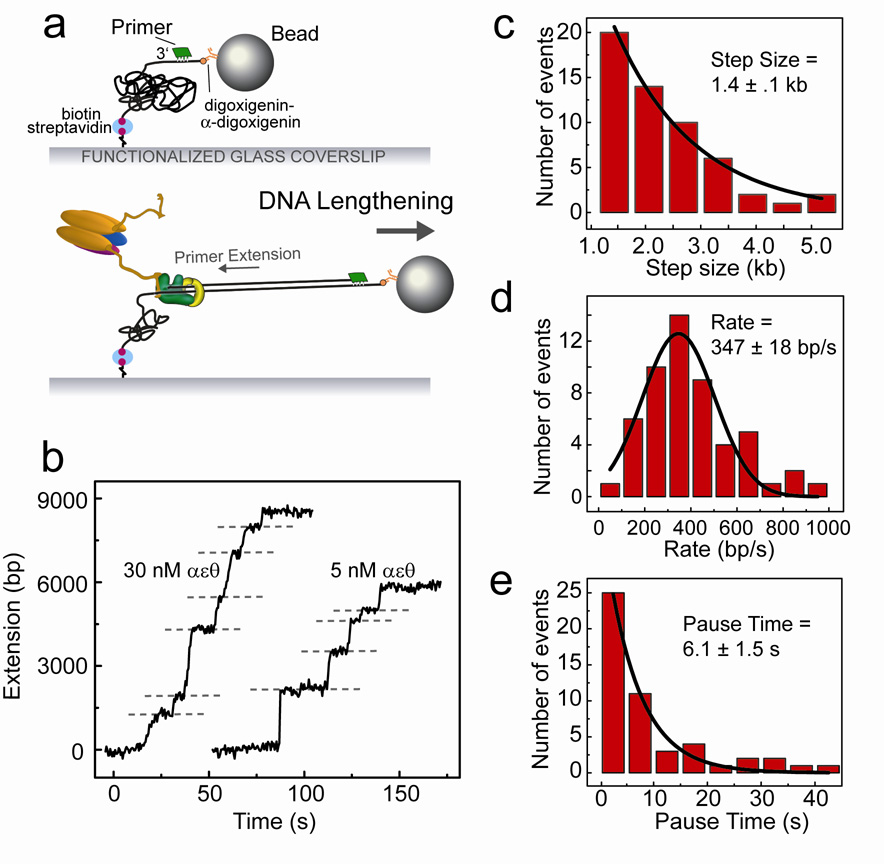
Figure 3. Primer extension by the Pol III holoenzyme.
Primer extension by the DNA polymerase III holoenzyme. (a) ssDNA is generated in the flow cell and an oligonucleotide primer is annealed (see text). Proteins are introduced with dNTPs and ATP, and primer-extension activity is measured as a lengthening of the tethered DNA. (b) Examples of lengthening traces, with extension at 30 nM αεθ on the left and 5 nM αεθ on the right. Dashed lines represent points where pauses in the DNA extension were identified. (c) Distribution of lengthening step sizes, fit with a single-exponential decay. (d) Distribution of rates, fit with a Gaussian distribution. (e) Distribution of lengths of pauses between lengthening steps, fit with a single-exponential decay. Data shown are from experiments performed with 30 nM Pol III holoenzyme.