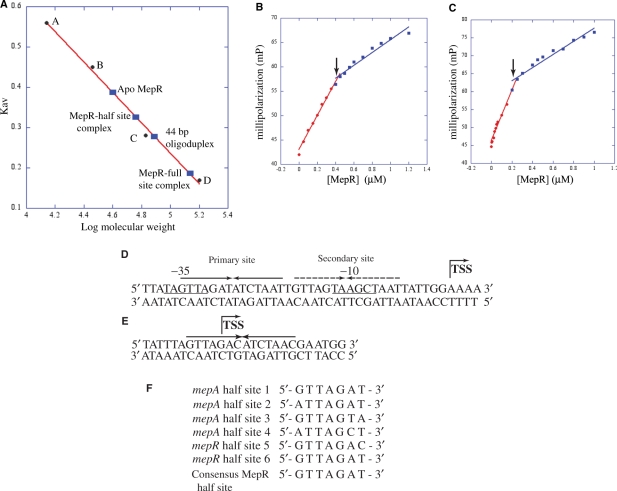
Figure 5.
Stoichiometry of MepR binding to the conserved pseudo palindromes located in the mepA and mepR promoters. (A) Size exclusion chromatography of MepR and MepR–DNA complexes. The linear fit of the elution values of four standard proteins [(A), ribonuclease A (Mr, 13.7 kDa); (B), carbonic anhydrase (Mr, 29 kDa); (C), bovine serum albumin (Mr, 66 kDa); (D), alcohol dehydrogenase (Mr, 150 kDa)] is shown as a red line. The Kav of MepR and MepR bound to the 44 and 27 bp high-affinity binding sites of the mepA promoter was plotted on the graph (solid blue rectangles) and labelled. (B and C) Fluorescence polarization-based determination of MepR–mepA-binding stoichiometry. (B) Binding to the 44 bp mepA operator site. (C) Binding to the 26 bp mepA operator site. The linear fit for the high affinity (specific) and low affinity (non-specific) binding are color coded in red and blue, respectively. The inflection points that denote the breaking point of high and low-affinity binding are indicated by black arrows. (D) The sequence of the MepR-binding sites on the mepA promoter. The 44-bp sequence comprising the MepR footprint of the mepA promoter is shown with the –10 and –35 hexamers of the promoter underlined and labelled. The primary binding site of MepR is indicated by solid inverted arrows, whereas the secondary site is shown as broken inverted arrows. The transcription start site (TSS) is marked by a bent arrow. (E) The sequence of the MepR-binding sites on the mepR promoter. The pseudo palindrome is shown as solid inverted arrows and the transcription start is indicated by the bent arrow. (F) Alignment of the heptanucleotide sequences of the MepR binding half sites of the mepA and mepR promoters. The half sites from the MepR primary (MepR sites 1 and 2) and secondary sites (sites 3 and 4) of the mepA operator and from the MepR-binding site of the mepR promoter (sites 5 and 6) are aligned from 5′→3′ direction and the consensus sequence derived from the sequence similarity amongst the six sequences is shown at the bottom of the alignment.