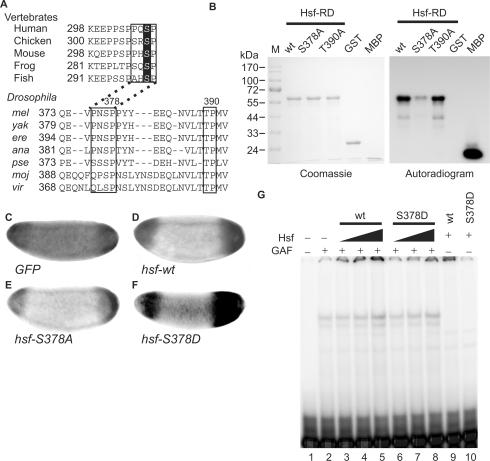
Figure 6.
Phosphorylation by Mapk converts Hsf from a repressor to an activator for tll expression. (A) Amino acid sequences of HSF1s in vertebrates were retrieved from Genbank and the regulatory domains of these HSF1s (HSF-RD) were aligned using the ClustalW program to reveal conserved amino acids. Putative Mapk recognition sequences are boxed and S307, a target site of Mapk in HSF1s, is highlighted. The same procedure was applied to Hsfs in Drosophila species including melanogaster (mel), yakuba (yak), erecta (ere), ananassae (ana), pseudoobscura (pse), mojavensis (moj) and virilis (vir). Amino acid sequences in boxes that contain two putative Mapk phosphorylation sites, S378 and T390, are highly conserved among Drosophila species. (B) In an in vitro phosphorylation assay, 100 pmol of GST-Hsf-RD and two mutant forms (GST-RD-S378A and T390A), as well as negative (GST) and positive (myelin basic protein, MBP) controls were used. After incubation with Mapk and γ[32P]-ATP, the proteins were separated in a 12% SDS gel and visualized using Coomassie blue staining, shown at the left. After the gel was dried, autoradiography was used to reveal the phosphorylated proteins, shown at the right. (C–F) tll expression patterns in late stage-4 embryos from mothers transheterozygous for hsf1 and TrlDHΔ34, determined by in situ hybridization, are shown and the anterior is arranged towards the left. Supplementation with UAS-hsf-wt (D), -S378A (E) and -S378D (F) driven by GAL4-GCN4 suppresses the expanded tll expression patterns. GFP expression was used as a negative control (C). (G) Hsf-S378D binding to the tor-RE is enhanced by GAF. The protein concentration of Hsf-S378D is 0.34 mg/ml. The Hsf, GAF and the probe were the same as those used in Figure 3A. GAF (4 μl) was mixed with various volumes of Hsf (wt) or Hsf-S378D (S378D) (2.5 μl in lanes 3 and 6; 5 μl in lanes 4 and 7; and 10 μl in lanes 5 and 8–10) and 32P-labeled probe. The DNA–protein complexes were separated in a 4% polyacrylamide gel and visualized using a PhosphoImager.